1ESL
 
 | |
3KMM
 
 | | Structure of human LCK kinase with a small molecule inhibitor | | Descriptor: | 3-(2,6-dichlorophenyl)-7-({4-[2-(diethylamino)ethoxy]phenyl}amino)-1-methyl-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Graves, B.J, Surgenor, A, Harris, W, Smith, I, Orchard, S, Flotow, H, Murray, E. | | Deposit date: | 2009-11-10 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human LCK kinase with a small molecule inhibitor
To be Published
|
|
2ILA
 
 | |
1HXB
 
 | | HIV-1 proteinase complexed with RO 31-8959 | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, HIV-1 PROTEASE | | Authors: | Graves, B.J, Hatada, M.H, Crowther, R.L. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel binding mode of highly potent HIV-proteinase inhibitors incorporating the (R)-hydroxyethylamine isostere.
J.Med.Chem., 34, 1991
|
|
1R36
 
 | | NMR-based structure of autoinhibited murine Ets-1 deltaN301 | | Descriptor: | C-ets-1 protein | | Authors: | Lee, G.M, Donaldson, L.W, Pufall, M.A, Kang, H.-S, Pot, I, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-09 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The Structural and Dynamic Basis of Ets-1 DNA Binding Autoinhibition
J.Biol.Chem., 280, 2005
|
|
1MDM
 
 | | INHIBITED FRAGMENT OF ETS-1 AND PAIRED DOMAIN OF PAX5 BOUND TO DNA | | Descriptor: | C-ETS-1 PROTEIN, PAIRED BOX PROTEIN PAX-5, PAX5/ETS BINDING SITE ON THE MB-1 PROMOTER | | Authors: | Garvie, C.W, Pufall, M.A, Graves, B.J, Wolberger, C. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STRUCTURAL ANALYSIS OF THE AUTOINHIBITION OF ETS-1 AND ITS ROLE IN PROTEIN PARTNERSHIPS
J.Biol.Chem., 277, 2002
|
|
1MD0
 
 | |
6DA1
 
 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.000127 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6DAT
 
 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35002637 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
2JUO
 
 | | GABPa OST domain | | Descriptor: | GA-binding protein alpha chain | | Authors: | Kang, H, Nelson, M.L, Mackereth, C.D, Schaerpf, M, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2007-08-31 | | Release date: | 2008-04-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and structural characterization of a CBP/p300-binding domain from the ETS family transcription factor GABP alpha
J.Mol.Biol., 377, 2008
|
|
2KMD
 
 | | Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to co-activator CBP | | Descriptor: | Protein C-ets-1 | | Authors: | Nelson, M.L, Kang, H, Lee, G.M, Blaszczak, A.G, Lau, D.K.W, McIntosh, L.P, Graves, B.J. | | Deposit date: | 2009-07-27 | | Release date: | 2010-05-05 | | Last modified: | 2012-03-21 | | Method: | SOLUTION NMR | | Cite: | Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LF7
 
 | |
2LF8
 
 | |
2MD5
 
 | | Structure of uninhibited ETV6 ETS domain | | Descriptor: | Transcription factor ETV6 | | Authors: | De, S, Mcintosh, L.P, Chan, A.C, Coyne, H.J, Okon, M, Graves, B.J, Murphy, M.E. | | Deposit date: | 2013-08-29 | | Release date: | 2013-12-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
4MHG
 
 | | Crystal structure of ETV6 bound to a specific DNA sequence | | Descriptor: | Complementary Specific 14 bp DNA, Specific 14 bp DNA, Transcription factor ETV6 | | Authors: | Chan, A.C, De, S, Coyne III, H.J, Okon, M, Murphy, M.E, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2013-08-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
8GJS
 
 | | Stapled Peptide ALRN-6924 Bound to MDMX | | Descriptor: | ACE-LEU-THR-PHE-ALA-GLU-TYR-TRP-ALA-GLN-LEU-DAL-ALA-ALA-ALA-ALA-ALA-DAL, Protein Mdm4 | | Authors: | Graves, B.J, Janson, C, Lukacs, C. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Sulanemadlin (ALRN-6924), the First Cell-Permeating, Stabilized alpha-Helical Peptide in Clinical Development.
J.Med.Chem., 66, 2023
|
|
4J3E
 
 | | The 1.9A crystal structure of humanized Xenopus Mdm2 with nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C.M, Kammlott, R.U, Crowther, R. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of RG7112: A Small-Molecule MDM2 Inhibitor in Clinical Development.
ACS Med Chem Lett, 4, 2013
|
|
4IPF
 
 | | The 1.7A crystal structure of humanized Xenopus MDM2 with RO5045337 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(4S,5R)-2-(4-tert-butyl-2-ethoxyphenyl)-4,5-bis(4-chlorophenyl)-4,5-dimethyl-4,5-dihydro-1H-imidazol-1-yl]{4-[3-(methylsulfonyl)propyl]piperazin-1-yl}methanone | | Authors: | Graves, B.J, Lukacs, C, Kammlott, R.U, Crowther, R. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | MDM2 Small-Molecule Antagonist RG7112 Activates p53 Signaling and Regresses Human Tumors in Preclinical Cancer Models.
Cancer Res., 73, 2013
|
|
4JRG
 
 | | The 1.9A crystal structure of humanized Xenopus MDM2 with RO5313109 - a pyrrolidine MDM2 inhibitor | | Descriptor: | (3R,4R,5S)-3-(3-chlorophenyl)-4-(4-chlorophenyl)-4-cyano-N-[(3S)-3,4-dihydroxybutyl]-5-(2,2-dimethylpropyl)-D-prolinamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Graves, B.J, Janson, C.A, Lukacs, C. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of RG7388, a Potent and Selective p53-MDM2 Inhibitor in Clinical Development.
J.Med.Chem., 56, 2013
|
|
4LWV
 
 | | The 2.3A Crystal Structure of Humanized Xenopus MDM2 with RO5545353 | | Descriptor: | (2S,3R,4R,5R)-N-(4-carbamoyl-2-methoxyphenyl)-2'-chloro-4-(3-chloro-2-fluorophenyl)-2-(2,2-dimethylpropyl)-5'-oxo-4',5'-dihydrospiro[pyrrolidine-3,6'-thieno[3,2-b]pyrrole]-5-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Janson, C.A. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of Potent and Orally Active p53-MDM2 Inhibitors RO5353 and RO2468 for Potential Clinical Development.
ACS MED.CHEM.LETT., 5, 2014
|
|
4LWT
 
 | | The 1.6A Crystal Structure of Humanized Xenopus MDM2 with RO5027344 | | Descriptor: | (3S)-3-[(3R)-1-acetylpiperidin-3-yl]-6-chloro-3-(3-chlorobenzyl)-1,3-dihydro-2H-indol-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Kammlott, U. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
4LWU
 
 | | The 1.14A Crystal Structure of Humanized Xenopus MDM2 with RO5499252 | | Descriptor: | (2'S,3R,4'S,5'R)-N-(4-carbamoylphenyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Janson, C.A. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
4N5T
 
 | |
3U15
 
 | | Structure of hDMX with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, Protein Mdm4, SULFATE ION | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VBG
 
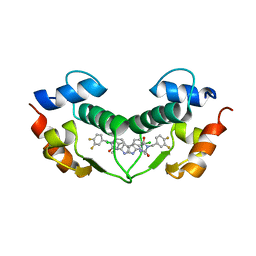 | | Structure of hDM2 with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2012-01-02 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
