4LGY
 
 | | Importance of Hydrophobic Cavities in Allosteric Regulation of Formylglycinamide Synthetase: Insight from Xenon Trapping and Statistical Coupling Analysis | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tanwar, A.S, Goyal, V.D, Choudhary, D, Panjikar, S, Anand, R. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Importance of hydrophobic cavities in allosteric regulation of formylglycinamide synthetase: insight from xenon trapping and statistical coupling analysis
Plos One, 8, 2013
|
|
4L78
 
 | | Xenon Trapping and Statistical Coupling Analysis Uncover Regions Important for Structure and Function of Multidomain Protein StPurL | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tanwar, A.S, Goyal, V.D, Choudhary, D, Panjikar, S, Anand, R. | | Deposit date: | 2013-06-13 | | Release date: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Importance of hydrophobic cavities in allosteric regulation of formylglycinamide synthetase: insight from xenon trapping and statistical coupling analysis
Plos One, 8, 2013
|
|
4MGH
 
 | | Importance of Hydrophobic Cavities in Allosteric Regulation of Formylglycinamide Synthetase: Insight from Xenon Trapping and Statistical Coupling Analysis | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tanwar, A.S, Goyal, V.D, Choudhary, D, Panjikar, S, Anand, R. | | Deposit date: | 2013-08-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Importance of hydrophobic cavities in allosteric regulation of formylglycinamide synthetase: insight from xenon trapping and statistical coupling analysis
Plos One, 8, 2013
|
|
6IGR
 
 | | Crystal structure of S9 peptidase (S514A mutant in inactive state) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Gaur, N.K, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IGP
 
 | | Crystal structure of S9 peptidase (inactive state)from Deinococcus radiodurans R1 in P212121 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IGQ
 
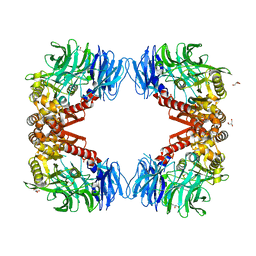 | | Crystal structure of inactive state of S9 peptidase from Deinococcus radiodurans R1 (PMSF treated) | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IKG
 
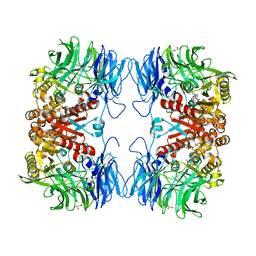 | | Crystal structure of substrate-bound S9 peptidase (S514A mutant) from Deinococcus radiodurans | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | Authors: | Yadav, P, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6A4R
 
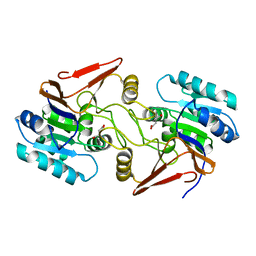 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | Descriptor: | ASPARTIC ACID, Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A4S
 
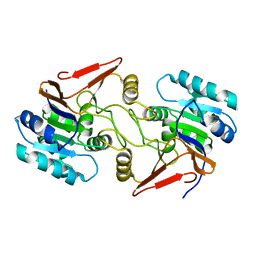 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | Descriptor: | Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A9U
 
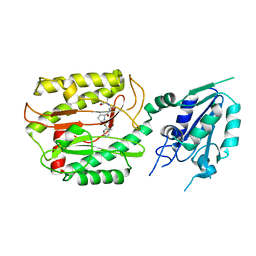 | | Crystal strcture of Icp55 from Saccharomyces cerevisiae bound to apstatin inhibitor | | Descriptor: | Intermediate cleaving peptidase 55, MANGANESE (II) ION, apstatin | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9V
 
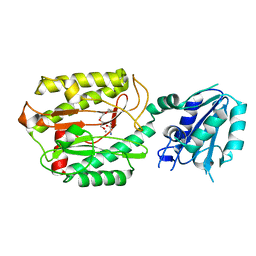 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9T
 
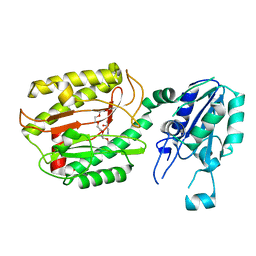 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 58 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
7CAY
 
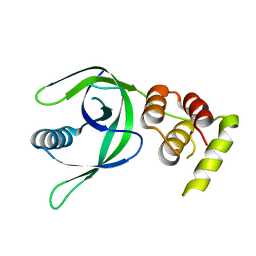 | | Crystal Structure of Lon N-terminal domain protein from Xanthomonas campestris | | Descriptor: | ATP-dependent protease | | Authors: | Singh, R, Sharma, B, Deshmukh, S, Kumar, A, Makde, R.D. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XCC3289 from Xanthomonas campestris: homology with the N-terminal substrate-binding domain of Lon peptidase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5GIQ
 
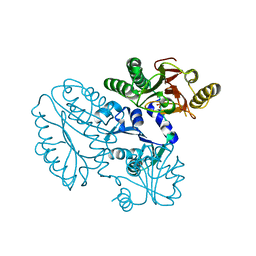 | | Xaa-Pro peptidase from Deinococcus radiodurans, Zinc bound | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, ZINC ION | | Authors: | Are, V.N, Singh, R, Kumar, A, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases.
Proteins, 2018
|
|
5GIV
 
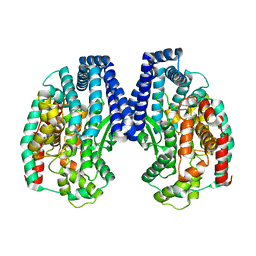 | | Crystal structure of M32 carboxypeptidase from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Carboxypeptidase 1, ZINC ION | | Authors: | Sharma, B, Singh, R, Yadav, P, Ghosh, B, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations
Biochim. Biophys. Acta, 1865, 2017
|
|
5CNX
 
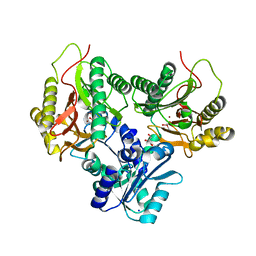 | | Crystal structure of Xaa-Pro aminopeptidase from Escherichia coli K12 | | Descriptor: | Aminopeptidase YpdF, CACODYLATE ION, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-18 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases
Proteins, 2018
|
|
5FCF
 
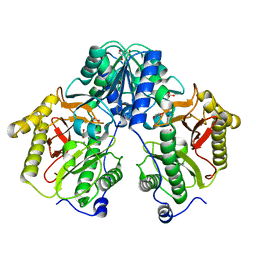 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Mn bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases.
Biochim. Biophys. Acta, 1865, 2017
|
|
5FCH
 
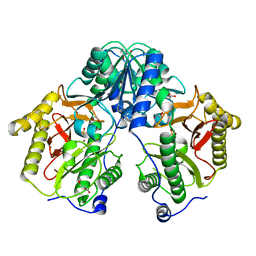 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Zn bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases
Biochim. Biophys. Acta, 1865, 2017
|
|
6A4T
 
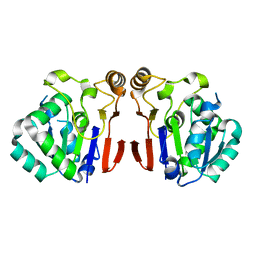 | | Crystal structure of Peptidase E from Deinococcus radiodurans R1 | | Descriptor: | Peptidase E | | Authors: | Yadav, P, Goyal, V.G, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic triad heterogeneity in S51 peptidase family: Structural basis for functional variability.
Proteins, 87, 2019
|
|
6IFG
 
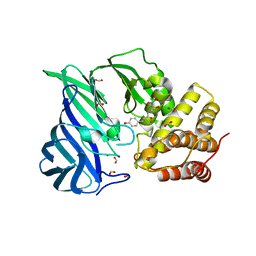 | | Crystal structure of M1 zinc metallopeptidase E323A mutant bound to Tyr-ser-ala substrate from Deinococcus radiodurans | | Descriptor: | FORMIC ACID, Tripeptides (TYR-SER-ALA), ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two-domain aminopeptidase of M1 family: Structural features for substrate binding and gating in absence of C-terminal domain.
J.Struct.Biol., 208, 2019
|
|
6IFF
 
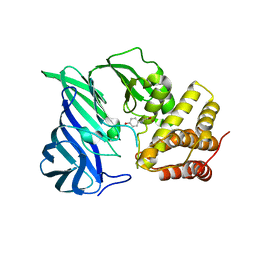 | | Crystal structure of M1 zinc metallopeptidase E323A mutant from Deinococcus radiodurans | | Descriptor: | SODIUM ION, TYROSINE, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Gaur, N.K, Makde, R.D. | | Deposit date: | 2018-09-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6IRU
 
 | |
6KP1
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-methionine amino acid | | Descriptor: | METHIONINE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KP0
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-arginine | | Descriptor: | ARGININE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KOZ
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323 mutant bound to L-Leucine amino acid | | Descriptor: | LEUCINE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
