3P24
 
 | | Structure of profragilysin-3 from Bacteroides fragilis | | Descriptor: | AZIDE ION, BFT-3, GLYCEROL, ... | | Authors: | Goulas, T, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2010-10-01 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure, function and latency regulation of a bacterial enterotoxin potentially derived from a mammalian adamalysin/ADAM xenolog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4YT9
 
 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) substrate-unbound. | | Descriptor: | GLYCEROL, Peptidylarginine deiminase, SODIUM ION | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
4YTB
 
 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) in complex with dipeptide Asp-Gln. | | Descriptor: | ASPARTIC ACID, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
4YTG
 
 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) mutant C351A in complex with dipeptide Met-Arg. | | Descriptor: | ARGININE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
5NCW
 
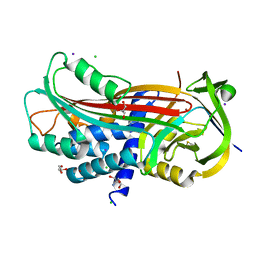 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin (V367K/K368A mutant). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCS
 
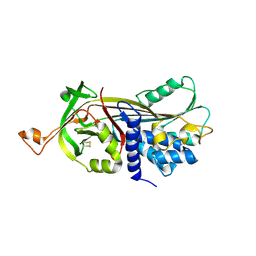 | | Structure of the native serpin-type proteinase inhibitor, miropin. | | Descriptor: | Serpin | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCT
 
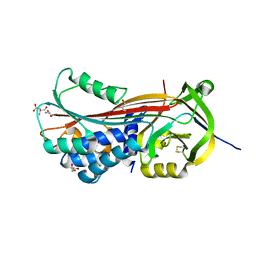 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin. | | Descriptor: | ASPARTIC ACID, GLYCEROL, SERINE, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCU
 
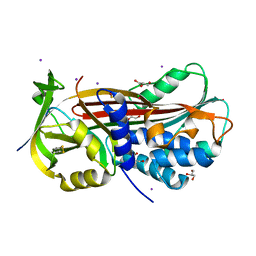 | | Structure of the subtilisin induced serpin-type proteinase inhibitor, miropin. | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5CX8
 
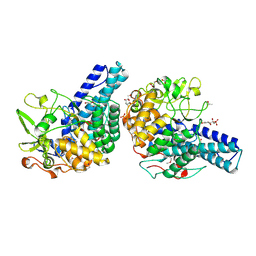 | | Structure of RagB, a major immunodominant virulence factor of Porphyromonas gingivalis. | | Descriptor: | 3-deoxy-5-O-phosphono-beta-D-ribofuranose, 3-deoxy-beta-D-glucopyranose, 6-O-phosphono-D-tagatose, ... | | Authors: | Goulas, T, Garcia-Ferrer, I, Hutcherson, J.A, Potempa, B.A, Potempa, J, Scott, D.A, Gomis-Ruth, F.X. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of RagB, a major immunodominant outer-membrane surface receptor antigen of Porphyromonas gingivalis.
Mol Oral Microbiol, 31, 2016
|
|
6TAV
 
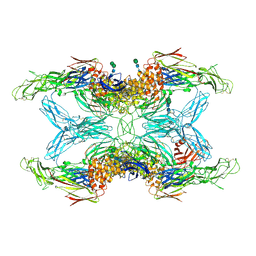 | | Crystal structure of endopeptidase-induced alpha2-macroglobulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Gomis-Ruth, F.X, Duquerroy, S, Trapani, S, Marrero, A, Goulas, T, Guevara, T, Andersen, G.A, Sottrup-Jensen, L, Navaza, J. | | Deposit date: | 2019-10-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Cryo-EM structures shows the mechanistic basis of pan-peptidase inhibition by human alpha2-macroglobulin
Proc.Natl.Acad.Sci.USA, 2022
|
|
8QB1
 
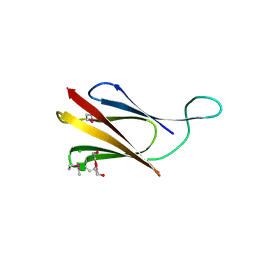 | | C-terminal domain of mirolase from Tannerella forsythia | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Mirolase, ... | | Authors: | Gomis-Ruth, F.X, Rodriguez-Banqueri, A, Mizgalska, D, Veillard, F, Goulas, T, Eckhard, U, Potempa, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional insights into the C-terminal signal domain of the Bacteroidetes type-IX secretion system
Open Biology, 2024
|
|
6ZA2
 
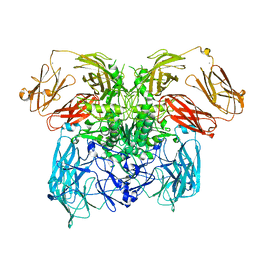 | | Crystal structure of dimeric latent PorU from Porphyromonas gingivalis | | Descriptor: | CALCIUM ION, Por secretion system protein porU | | Authors: | Gomis-Ruth, F.X, Goulas, T, Guevara, T, Rodriguez-Banqueri, A, Potempa, J. | | Deposit date: | 2020-06-04 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Intermolecular latency regulates the essential C-terminal signal peptidase and sortase of the Porphyromonas gingivalis type-IX secretion system.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8B2Q
 
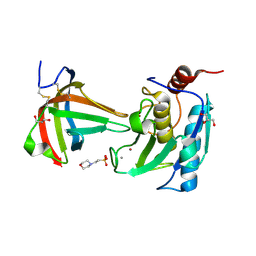 | | Matrix-metallopeptidase inhibitor Potempin A (PotA) from Tannerella forsythia in complex with T. forsythia karilysin. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Potempa, J, Ksiazek, M, Goulas, T, Cuppari, A, Rodriguez-Banqueri, A, Arolas, J.L, Lopez-Pelegrin, M, Garcia-Ferrer, I, Guevara, T. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Chem Sci, 14, 2023
|
|
8B2N
 
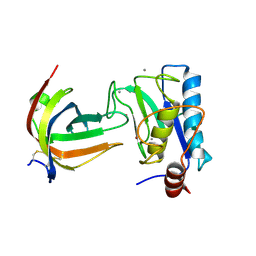 | | Potempin A (PotA) from Tannerella forsythia in complex with the catalytic domain of human MMP-12 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, Tannerella forsythia potempin A (PotA), ... | | Authors: | Potempa, J, Ksiazek, M, Goulas, T, Cuppari, A, Rodriguez-Banqueri, A, Arolas, J.L, Lopez-Pelegrin, M, Garcia-Ferrer, I, Guevara, T. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Chem Sci, 14, 2023
|
|
8B2M
 
 | | Matrix-metallopeptidase inhibitor Potempin A (PotA) from Tannerella forsythia | | Descriptor: | NICKEL (II) ION, Tannerella forsythia Potempin A (PotA) | | Authors: | Potempa, J, Ksiazek, M, Goulas, T, Cuppari, A, Rodriguez-Banqueri, A, Arolas, J.L, Lopez-Pelegrin, M, Garcia-Ferrer, I, Guevara, T. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Chem Sci, 14, 2023
|
|
6I0X
 
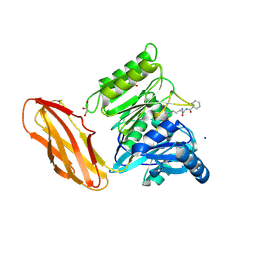 | | Porphyromonas gingivalis peptidylarginine deminase (PPAD) mutant G231N/E232T/N235D in complex with Cl-amidine. | | Descriptor: | GLYCEROL, N-[(1S)-1-(AMINOCARBONYL)-4-(ETHANIMIDOYLAMINO)BUTYL]BENZAMIDE, Peptidylarginine deiminase, ... | | Authors: | Gomis-Ruth, F.X, Goulas, T, Sola, M, Potempa, J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, function, and inhibition of a genomic/clinical variant of Porphyromonas gingivalis peptidylarginine deiminase.
Protein Sci., 28, 2019
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
5M11
 
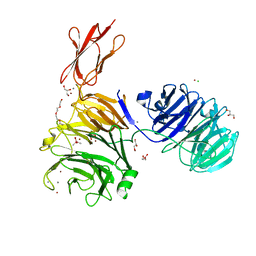 | | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lasica, A.M, Goulas, T, Mizgalska, D, Zhou, X, Ksiazek, M, Madej, M, Guo, Y, Guevara, T, Nowak, M, Potempa, B, Goel, A, Sztukowska, M, Prabhakar, A.T, Bzowska, M, Widziolek, M, Thogersen, I.B, Enghild, J.J, Simonian, M, Kulczyk, A.W, Nguyen, K.-A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis.
Sci Rep, 6, 2016
|
|
5MUN
 
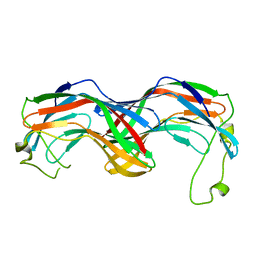 | | Structural insight into zymogenic latency of gingipain K from Porphyromonas gingivalis. | | Descriptor: | AZIDE ION, Lys-gingipain W83 | | Authors: | Pomowski, A, Uson, I, Nowakovska, Z, Veillard, F, Sztukowska, M.N, Guevara, T, Goulas, T, Mizgalska, D, Nowak, M, Potempa, B, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
6HT9
 
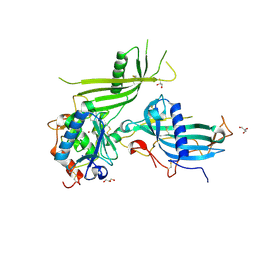 | | Mouse fetuin-B in complex with crayfish astacin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Astacin, ... | | Authors: | Gomis-Ruth, F.X, Goulas, T, Guevara, T, Cuppari, A. | | Deposit date: | 2018-10-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition.
Iucrj, 6, 2019
|
|
5NV6
 
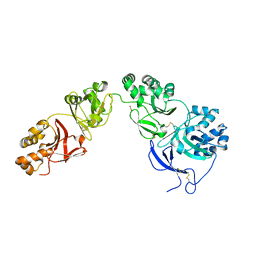 | | Structure of human transforming growth factor beta-induced protein (TGFBIp). | | Descriptor: | ACETATE ION, Transforming growth factor-beta-induced protein ig-h3 | | Authors: | Garcia-Castellanos, R, Nielsen, S.N, Runager, K, Thogersen, B.I, Goulas, T, Enghild, J.J, Gomis-Ruth, F.X. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural and Functional Implications of Human Transforming Growth Factor beta-Induced Protein, TGFBIp, in Corneal Dystrophies.
Structure, 25, 2017
|
|
7O7N
 
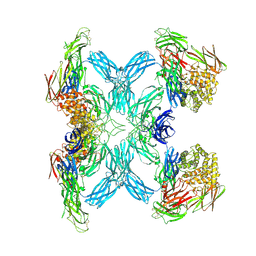 | | (h-alpha2M)4 semiactivated I state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7O7R
 
 | | (h-alpha2M)4 plasmin-activated I state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7O7P
 
 | | (h-alpha2M)4 activated state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7O7Q
 
 | | (h-alpha2M)4 trypsin-activated state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
