6XE0
 
 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
1DKZ
 
 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 1 NATIVE CRYSTALS | | Descriptor: | SUBSTRATE BINDING DOMAIN OF DNAK, SUBSTRATE PEPTIDE (7 RESIDUES) | | Authors: | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
1DKX
 
 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 1 SELENOMETHIONYL CRYSTALS | | Descriptor: | SUBSTRATE BINDING DOMAIN OF DNAK, SUBSTRATE PEPTIDE (7 RESIDUES) | | Authors: | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
1DKY
 
 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 2 NATIVE CRYSTALS | | Descriptor: | DNAK, PEPTIDE SUBSTRATE | | Authors: | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
6ER0
 
 | | 6th KOW domain of human hSpt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Hahn, L, Schweimer, K, Roesch, P, Woehrl, B.M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-31 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure and nucleic acid binding properties of KOW domains 4 and 6-7 of human transcription elongation factor DSIF.
Sci Rep, 8, 2018
|
|
6EQY
 
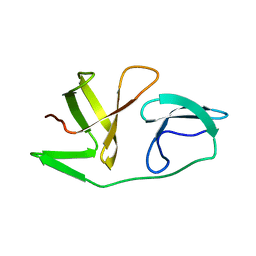 | | 4th KOW domain of human hSpt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Zuber, P.K, Schweimer, K, Roesch, P, Woehrl, B.M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-17 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure and nucleic acid binding properties of KOW domains 4 and 6-7 of human transcription elongation factor DSIF.
Sci Rep, 8, 2018
|
|
3IMQ
 
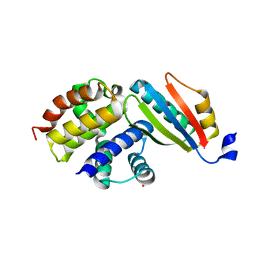 | | Crystal structure of the NusB101-S10(delta loop) complex | | Descriptor: | 30S ribosomal protein S10, N utilization substance protein B, POTASSIUM ION | | Authors: | Luo, X, Wahl, M.C. | | Deposit date: | 2009-08-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination.
Nucleic Acids Res., 38, 2010
|
|
2XHA
 
 | |
2XHC
 
 | |
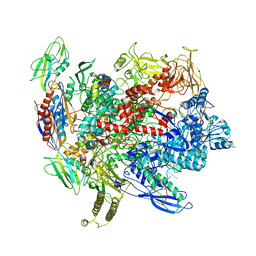
6ALG
 
 | |
6ALF
 
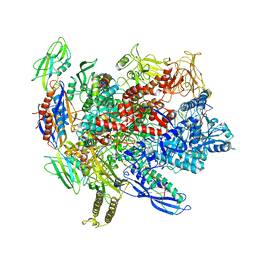 | | CryoEM structure of crosslinked E.coli RNA polymerase elongation complex | | Descriptor: | DNA (29-MER), DNA (5'-D(*GP*GP*GP*CP*TP*AP*AP*TP*GP*AP*CP*GP*GP*CP*GP*AP*AP*TP*AP*CP*CP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Darst, S.A. | | Deposit date: | 2017-08-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription arrest by coliphage HK022 Nun in anEscherichia coliRNA polymerase elongation complex.
Elife, 6, 2017
|
|
6ALH
 
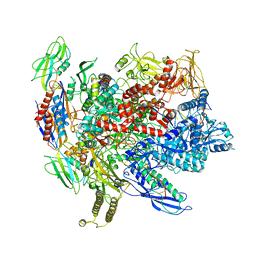 | | CryoEM structure of E.coli RNA polymerase elongation complex | | Descriptor: | DNA (29-MER), DNA (5'-D(*GP*GP*GP*CP*TP*AP*AP*TP*GP*AP*CP*GP*GP*CP*GP*AP*AP*TP*AP*CP*CP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Darst, S.A. | | Deposit date: | 2017-08-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of transcription arrest by coliphage HK022 Nun in anEscherichia coliRNA polymerase elongation complex.
Elife, 6, 2017
|
|
2KVQ
 
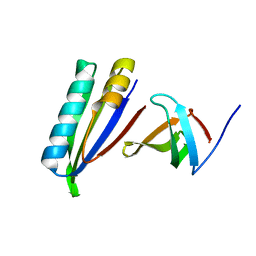 | |
2LQ8
 
 | | Domain interaction in Thermotoga maritima NusG | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Droegemueller, J, Stegmann, C, Burmann, B, Roesch, P, Wahl, M.C, Schweimer, K. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Autoinhibited State in the Structure of Thermotoga maritima NusG.
Structure, 21, 2013
|
|
3D3C
 
 | |
3D3B
 
 | |
