5J04
 
 | |
4ROS
 
 | |
4ROP
 
 | |
5UGR
 
 | | Malyl-CoA lyase from Methylobacterium extorquens | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Malyl-CoA lyase/beta-methylmalyl-CoA lyase, ... | | Authors: | Gonzalez, J.M. | | Deposit date: | 2017-01-09 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of Methylobacterium extorquens malyl-CoA lyase: CoA-substrate binding correlates with domain shift.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5UJK
 
 | |
5ULV
 
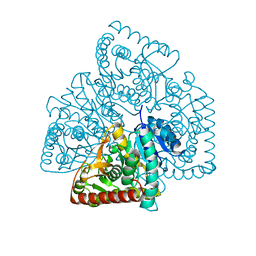 | | Malate dehydrogenase from Methylobacterium extorquens | | Descriptor: | CALCIUM ION, Malate dehydrogenase | | Authors: | Gonzalez, J.M. | | Deposit date: | 2017-01-25 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational changes on substrate binding revealed by structures of Methylobacterium extorquens malate dehydrogenase.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3I11
 
 | |
3I0V
 
 | |
3I14
 
 | |
3I15
 
 | |
3I13
 
 | |
3RZY
 
 | |
3P6G
 
 | |
3P6D
 
 | |
3P6F
 
 | |
3P6C
 
 | |
3P6E
 
 | |
3P6H
 
 | |
4NQ5
 
 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 complexed with compound CS319 | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase 2, POTASSIUM ION, ... | | Authors: | Gonzalez, J.M, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Inhibition of metallo-lactamases with bicyclic compounds
TO BE PUBLISHED
|
|
4NQ4
 
 | |
4NQ6
 
 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 complexed with compound L-CS319 | | Descriptor: | (3S,5S,7aR)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase 2, POTASSIUM ION, ... | | Authors: | Gonzalez, J.M, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6BNX
 
 | | Crystal structure of V278E-glyoxalase I mutant from Zea mays in space group P6(3) | | Descriptor: | COBALT (II) ION, Lactoylglutathione lyase | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
6BNZ
 
 | | Crystal structure of E144Q-glyoxalase I mutant from Zea mays in space group P4(1)2(1)2 | | Descriptor: | COBALT (II) ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
6BNN
 
 | | Crystal structure of V278E-glyoxalase I mutant from Zea mays in space group P4(1)2(1)2 | | Descriptor: | COBALT (II) ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
2NZE
 
 | | Structure of beta-lactamase II from Bacillus cereus. R121H, C221S double mutant. Space group P3121. | | Descriptor: | ACETIC ACID, Beta-lactamase II, GLYCEROL, ... | | Authors: | Medrano Martin, F.J, Vila, A.J, Gonzalez, J.M. | | Deposit date: | 2006-11-23 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Zn2 position in metallo-beta-lactamases is critical for activity: a study on chimeric metal sites on a conserved protein scaffold.
J.Mol.Biol., 373, 2007
|
|
