219D
 
 | |
1E68
 
 | | Solution structure of bacteriocin AS-48 | | Descriptor: | AS-48 PROTEIN | | Authors: | Gonzalez, C, Langdon, G, Bruix, M, Galvez, A, Valdivia, E, Maqueda, M, Rico, M. | | Deposit date: | 2000-08-09 | | Release date: | 2000-10-25 | | Last modified: | 2011-08-03 | | Method: | SOLUTION NMR | | Cite: | Bacteriocin as-48, a Microbial Cyclic Polypeptide Structurally and Functionally Related to Mammalian Nk-Lysin
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1H20
 
 | | Solution structure of the potato carboxypeptidase inhibitor | | Descriptor: | METALLOCARBOXYPEPTIDASE INHIBITOR | | Authors: | Gonzalez, C, Neira, J.L, Ventura, S, Bronsoms, S, Aviles, F.X, Rico, M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-05-09 | | Last modified: | 2011-12-28 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Potato Carboxypeptidase Inhibitor by 1H and 15N NMR.
Proteins, 50, 2003
|
|
2KP4
 
 | | Structure of 2'F-ANA/RNA hybrid duplex | | Descriptor: | DNA (5'-D(*(GFL)P*(CFL)P*(TAF)P*(A5L)P*(TAF)P*(A5L)P*(A5L)P*(TAF)P*(GFL)P*(GFL))-3'), RNA (5'-R(*CP*CP*AP*UP*UP*AP*UP*AP*GP*C)-3') | | Authors: | Gonzalez, C, Martin-Pintado, N, Watts, J, Gomez-Pinto, I, Dhama, M, Orozco, M, Schwartzentruber, J, Portella, G. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Differential stability of 2'F-ANA*RNA and ANA*RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility.
Nucleic Acids Res., 38, 2010
|
|
2KP3
 
 | | Structure of ANA-RNA hybrid duplex | | Descriptor: | RNA (5'-R(*(GAO)P*(CAR)P*(UAR)P*(A5O)P*(UAR)P*(A5O)P*(A5O)P*(UAR)P*(GAO)P*(GAO))-3'), RNA (5'-R(*CP*CP*AP*UP*UP*AP*UP*AP*GP*C)-3') | | Authors: | Gonzalez, C, Martn-Pintado, N, Watts, J, Gomez-Pinto, I, Dhama, M, Orozco, M, Schwartzentruber, J, Portella, G. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Differential stability of 2'F-ANA*RNA and ANA*RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility.
Nucleic Acids Res., 38, 2010
|
|
2AAS
 
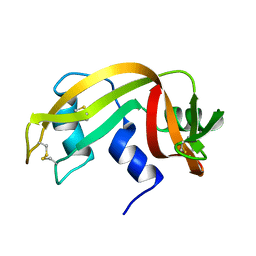 | | HIGH-RESOLUTION THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE A IN SOLUTION BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | RIBONUCLEASE A | | Authors: | Santoro, J, Gonzalez, C, Bruix, M, Neira, J.L, Nieto, J.L, Herranz, J, Rico, M. | | Deposit date: | 1992-11-20 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 229, 1993
|
|
2CMN
 
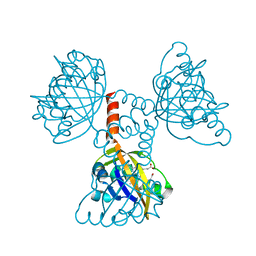 | | A Proximal Arginine Residue in the Switching Mechanism of the FixL Oxygen Sensor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gilles-Gonzalez, M.-A, Caceres, A.I, Silva Sousa, E.H, Tomchick, D.R, Brautigam, C.A, Gonzalez, C, Machius, M. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Proximal Arginine R206 Participates in Switching of the Bradyrhizobium Japonicum Fixl Oxygen Sensor
J.Mol.Biol., 360, 2006
|
|
8OFC
 
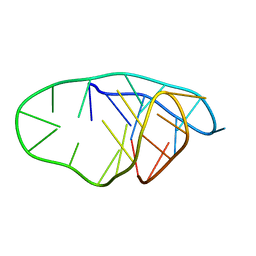 | | Structure of an i-motif domain with the cytosine analog 1,3-diaza-2-oxophenoxacione (tC) at neutral pH | | Descriptor: | DNA (5'-D(*CP*(YCO)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*(DNR)P*GP*T)-3') | | Authors: | Mir, B, Serrano-Chacon, I, Terrazas, M, Gandioso, A, Garavis, M, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2023-03-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Site-specific incorporation of a fluorescent nucleobase analog enhances i-motif stability and allows monitoring of i-motif folding inside cells.
Nucleic Acids Res., 2024
|
|
8PWR
 
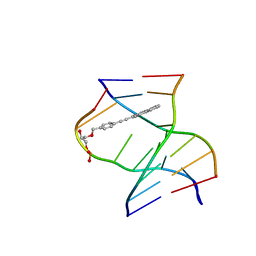 | | TINA-conjugated antiparallel DNA triplex | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*GP*GP*TP*GP*(J32)P*GP*T)-3') | | Authors: | Garavis, M, Edwards, P.J.B, Serrano-Chacon, I, Doluca, O, Filichev, V.V, Gonzalez, C. | | Deposit date: | 2023-07-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-27 | | Method: | SOLUTION NMR | | Cite: | Understanding intercalative modulation of G-rich sequence folding: solution structure of a TINA-conjugated antiparallel DNA triplex.
Nucleic Acids Res., 52, 2024
|
|
1N96
 
 | | DIMERIC SOLUTION STRUCTURE OF THE CYCLIC OCTAMER CD(CGCTCATT) | | Descriptor: | CYCLIC OLIGONUCLEOTIDE D(CGCTCATT) | | Authors: | Escaja, N, Gelpi, J.L, Orozco, M, Rico, M, Pedroso, E, Gonzalez, C. | | Deposit date: | 2002-11-22 | | Release date: | 2003-05-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Four-stranded DNA structure stabilized by a novel G:C:A:T tetrad.
J.Am.Chem.Soc., 125, 2003
|
|
1TRL
 
 | | NMR SOLUTION STRUCTURE OF THE C-TERMINAL FRAGMENT 255-316 OF THERMOLYSIN: A DIMER FORMED BY SUBUNITS HAVING THE NATIVE STRUCTURE | | Descriptor: | THERMOLYSIN FRAGMENT 255 - 316 | | Authors: | Rico, M, Jimenez, M.A, Gonzalez, C, De Filippis, V, Fontana, A. | | Deposit date: | 1994-09-02 | | Release date: | 1995-02-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the C-terminal fragment 255-316 of thermolysin: a dimer formed by subunits having the native structure.
Biochemistry, 33, 1994
|
|
2WAD
 
 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN 5204) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2B, ZINC ION | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
7NEJ
 
 | | Solution structure of DNA:RNA hybrid duplex | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*AP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*UP*GP*UP*AP*G)-3') | | Authors: | Cabrero, C, Martin-Pintado, N, Mazzini, S, Gargallo, R, Eritja, R, Avino, A, Gonzalez, C. | | Deposit date: | 2021-02-04 | | Release date: | 2021-04-07 | | Method: | SOLUTION NMR | | Cite: | Structural Effects of Incorporation of 2 -Deoxy-2 2 -difluorodeoxycytidine (Gemcitabine) in A- and B-form Duplexes.
Chemistry, 2021
|
|
7NBL
 
 | | Solution structure of DNA:RNA hybrid containing a 2'-deoxy-2'2'-difluorodeoxycytidine (gemcitabine) modification | | Descriptor: | DNA (5'-D(*CP*TP*AP*(FFC)P*AP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*UP*GP*UP*AP*G)-3') | | Authors: | Cabrero, C, Martin-Pintado, N, Mazzini, S, Gargallo, R, Eritja, R, Avino, A, Gonzalez, C. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Method: | SOLUTION NMR | | Cite: | Structural Effects of Incorporation of 2 -Deoxy-2 2 -difluorodeoxycytidine (Gemcitabine) in A- and B-form Duplexes.
Chemistry, 2021
|
|
7NBK
 
 | | Solution structure of DNA duplex containing a 2'-deoxy-2'2'-difluorodeoxycytidine (gemcitabine) modification | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*(FFC)P*G)-3') | | Authors: | Cabrero, C, Martin-Pintado, N, Mazzini, S, Gargallo, R, Eritja, R, Avino, A, Gonzalez, C. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Method: | SOLUTION NMR | | Cite: | Structural Effects of Incorporation of 2 -Deoxy-2 2 -difluorodeoxycytidine (Gemcitabine) in A- and B-form Duplexes.
Chemistry, 2021
|
|
7NBP
 
 | | Solution structure of DNA duplex containing a 7,8-dihydro-8-oxo-1,N6-ethenoadenine base modification that induces exclusively A->T transversions in Escherichia coli | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*(EDA)P*CP*AP*TP*G)-3') | | Authors: | Aralov, A.V, Gubina, N, Cabrero, C, Tsvetkov, V.B, Turaev, A.V, Fedeles, B.I, Croy, R.G, Isaakova, E.A, Melnik, D, Dukova, S, Khrulev, A.A, Varizhuk, A.M, Gonzalez, C, Zatsepin, T.S, Essigmann, J.M. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-04-20 | | Method: | SOLUTION NMR | | Cite: | 7,8-Dihydro-8-oxo-1,N6-ethenoadenine: an exclusively Hoogsteen-paired thymine mimic in DNA that induces A→T transversions in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
1GL8
 
 | | Solution structure of thioredoxin m from spinach, oxidized form | | Descriptor: | THIOREDOXIN | | Authors: | Neira, J.L, Gonzalez, C, Toiron, C, De-Prat-gay, G, Rico, M. | | Deposit date: | 2001-08-30 | | Release date: | 2001-12-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Solution Structure and Stability of Thioredoxin M from Spinach.
Biochemistry, 40, 2001
|
|
1EU2
 
 | |
1EU6
 
 | |
3GWH
 
 | | Crystallographic Ab Initio protein solution far below atomic resolution | | Descriptor: | PHOSPHATE ION, Transcriptional antiterminator (BglG family) | | Authors: | Rodriguez, D.D, Grosse, C, Himmel, S, Gonzalez, C, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic ab initio protein structure solution below atomic resolution
Nat.Methods, 6, 2009
|
|
1RSB
 
 | |
2KSS
 
 | | NMR structure of Myxococcus xanthus antirepressor CarS1 | | Descriptor: | Carotenogenesis protein carS | | Authors: | Jimenez, M, Gonzalez, C, Padmanabhan, S, Leon, E, Navarro-Aviles, G, Elias-Arnanz, M. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix.
Nucleic Acids Res., 38, 2010
|
|
2WAE
 
 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN 5204) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2B, ZINC ION | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2WAF
 
 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN R6) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 2B | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2N89
 
 | | Tetrameric i-motif structure of dT-dC-dC-CFL-CFL-dC at acidic pH | | Descriptor: | DNA (5'-D(*TP*CP*CP*(CFL)P*(CFL)P*C)-3') | | Authors: | Abou-Assi, H, Harkness, R.W, Martin-Pintado, N, Wilds, C.J, Campos-Olivas, R, Mittermaier, A.K, Gonzalez, C, Damha, M.J. | | Deposit date: | 2015-10-09 | | Release date: | 2016-08-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Stabilization of i-motif structures by 2'-beta-fluorination of DNA.
Nucleic Acids Res., 44, 2016
|
|
