8END
 
 | |
8ENB
 
 | |
8ENF
 
 | |
8FM7
 
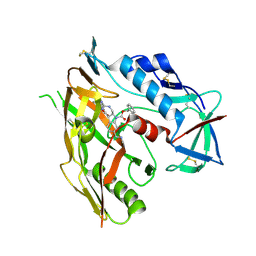 | | HIV-1 gp120 complex with CJF-III-192 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, benzyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM2
 
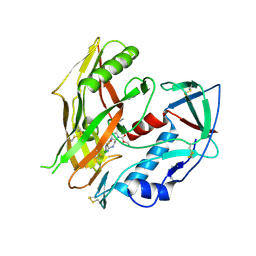 | | HIV-1 gp120 complex with CJF-III-289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, IMIDAZOLE, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM3
 
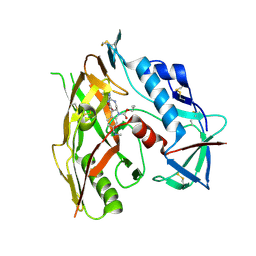 | | HIV-1 gp120 complex with CJF-III-288 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, propyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM8
 
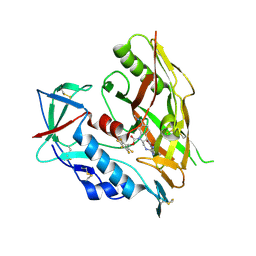 | | HIV-1 gp120 complex with CJF-IV-046 | | Descriptor: | (pentafluorophenyl)methyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FLY
 
 | | HIV-1 gp120 complex with BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-5-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FLZ
 
 | | HIV-1 gp120 complex with CJF-III-049-S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3S)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM4
 
 | | HIV-1 gp120 complex with CJF-IV-047 | | Descriptor: | 2,2,2-trifluoroethyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120 | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM5
 
 | | HIV-1 gp120 complex with DY-III-065 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,3,3-trifluoropropyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate, Envelope glycoprotein gp120 | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM0
 
 | | HIV-1 gp120 complex with CJF-III-214 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, methyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Indoline CD4-mimetic compounds mediate potent and broad HIV-1 inhibition and sensitization to antibody-dependent cellular cytotoxicity.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TJP
 
 | | HIV-1 gp120 complex with CJF-II-195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, IMIDAZOLE, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural and Functional Characterization of Indane-Core CD4-Mimetic Compounds Substituted with Heterocyclic Amines
Acs Medicinal Chemistry Letters, 14, 2023
|
|
7TJO
 
 | | HIV-1 gp120 complex with CJF-II-197-S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, IMIDAZOLE, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural and Functional Characterization of Indane-Core CD4-Mimetic Compounds Substituted with Heterocyclic Amines
ACS Medicinal Chemistry Letters, 14, 2023
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
8H0N
 
 | | Crystal structure of the human METTL1-WDR4 complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Jin, X.H, Guan, Z.Y, Gong, Z, Zhang, D.L. | | Deposit date: | 2022-09-30 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into how WDR4 promotes the tRNA N7-methylguanosine methyltransferase activity of METTL1.
Cell Discov, 9, 2023
|
|
2N2K
 
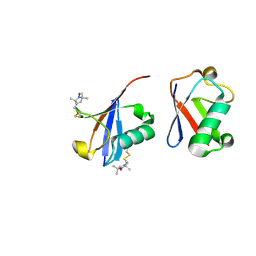 | | Ensemble structure of the closed state of Lys63-linked diubiquitin in the absence of a ligand | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ubiquitin | | Authors: | Liu, Z, Gong, Z, Tang, C. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-08 | | Last modified: | 2015-09-23 | | Method: | SOLUTION NMR | | Cite: | Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition.
Elife, 4, 2015
|
|
2D2A
 
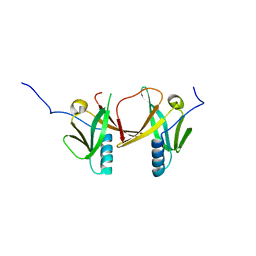 | | Crystal Structure of Escherichia coli SufA Involved in Biosynthesis of Iron-sulfur Clusters | | Descriptor: | SufA protein | | Authors: | Wada, K, Hasegawa, Y, Gong, Z, Minami, Y, Fukuyama, K, Takahashi, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: Implications for a functional dimer
Febs Lett., 579, 2005
|
|
5XK4
 
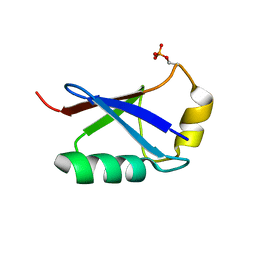 | | Retracted state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Dong, X, Gong, Z, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YMY
 
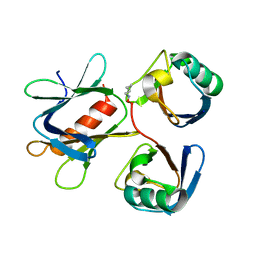 | | The structure of the complex between Rpn13 and K48-diUb | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Liu, Z, Dong, X, Gong, Z, Yi, H.W, Liu, K, Yang, J, Zhang, W.P, Tang, C. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-13 | | Last modified: | 2019-04-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13.
Cell Discov, 5, 2019
|
|
5YZ9
 
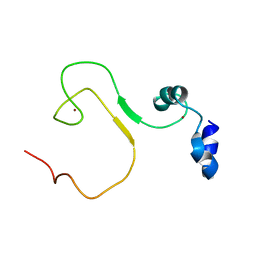 | | zinc finger domain of METTL3-METTL14 N6-methyladenosine methyltransferase | | Descriptor: | N6-adenosine-methyltransferase catalytic subunit, ZINC ION | | Authors: | Dong, X, Tang, C, Gong, Z, Yin, P, Huang, J.B. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-28 | | Last modified: | 2019-03-27 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase.
Protein Cell, 10, 2019
|
|
5B6D
 
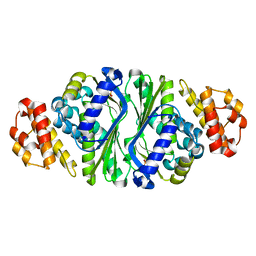 | |
5B6E
 
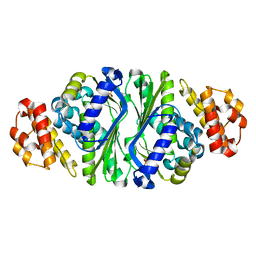 | |
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
