8IRK
 
 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-18 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Mycobacterial ergothioneine-biosynthesis C-S lyase EgtE
To Be Published
|
|
7EY2
 
 | |
7EY1
 
 | | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-xylosidase/beta-D-glucosidase, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose
To Be Published
|
|
6JBS
 
 | | Bifunctional xylosidase/glucosidase LXYL | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2019-01-26 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of beta-glycosidase LXYL-P1-2 reveals the product binding state of GH3 family and a specific pocket for Taxol recognition.
Commun Biol, 3, 2020
|
|
6KJ0
 
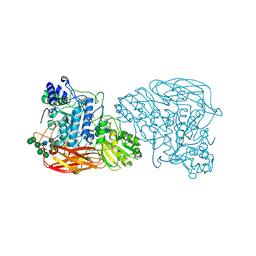 | | Bifunctional xylosidase/glucosidase LXYL mutant E529Q C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-xylosidase/beta-D-glucosidase, Deacetyltaxol, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2019-07-20 | | Release date: | 2020-02-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of beta-glycosidase LXYL-P1-2 reveals the product binding state of GH3 family and a specific pocket for Taxol recognition.
Commun Biol, 3, 2020
|
|
3NTI
 
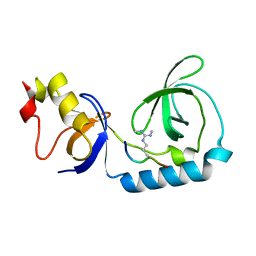 | | Crystal structure of Tudor and Aubergine [R15(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
3NTK
 
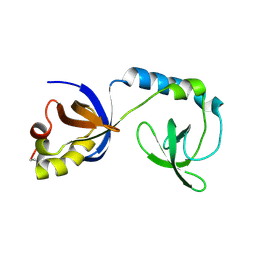 | | Crystal structure of Tudor | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
3NTH
 
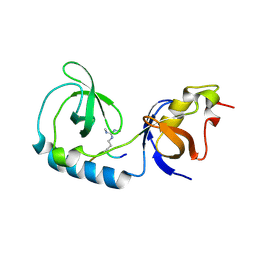 | | Crystal structure of Tudor and Aubergine [R13(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
4ORD
 
 | |
8IS0
 
 | | Carbon Sulfoxide lyase - Y106F | | Descriptor: | 2-AMINO-ACRYLIC ACID, PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRZ
 
 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRY
 
 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8H7V
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H81
 
 | | Trans-3/4-proline-hydroxylase H11 with 4-Hydroxyl-proline | | Descriptor: | 4-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7T
 
 | | Trans-3/4-proline-hydroxylase H11 apo structure | | Descriptor: | CHLORIDE ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H85
 
 | | Trans-3/4-proline-hydroxylase H11 with 3-hydroxyl-proline | | Descriptor: | 3-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7Y
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, PROLINE, ... | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7YO7
 
 | | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose, 5 seconds | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-1)-alpha-D-mannopyranose-(3-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose, ... | | Authors: | Yang, L.Y. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose
To Be Published
|
|
8G05
 
 | | Cryo-EM structure of an orphan GPCR-Gi protein signaling complex | | Descriptor: | 6-(octylamino)pyrimidine-2,4(3H,5H)-dione, CHOLESTEROL, G-protein coupled receptor 84, ... | | Authors: | Zhang, X, Wang, Y.J, Li, X, Liu, G.B, Gong, W.M, Zhang, C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Pro-phagocytic function and structural basis of GPR84 signaling.
Nat Commun, 14, 2023
|
|
5ED9
 
 | | Crystal structure of CC1 of mouse SUN2 | | Descriptor: | SUN domain-containing protein 2 | | Authors: | Nie, S, Ke, H.M, Gao, F, Ren, J.Q, Wang, M.Z, Huo, L, Gong, W.M, Feng, W. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators
Structure, 24, 2016
|
|
5ED8
 
 | | Crystal structure of CC2-SUN of mouse SUN2 | | Descriptor: | MAGNESIUM ION, MKIAA0668 protein | | Authors: | Nie, S, Ke, H.M, Gao, F, Ren, J.Q, Wang, M.Z, Huo, L, Gong, W.M, Feng, W. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators
Structure, 24, 2016
|
|
4KK1
 
 | | Crystal Structure of TSC1 core domain from S. pombe | | Descriptor: | Tuberous sclerosis 1 protein homolog | | Authors: | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-17 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
4KK0
 
 | | Crystal Structure of TSC1 core domain from S. pombe | | Descriptor: | Tuberous sclerosis 1 protein homolog | | Authors: | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-17 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
6AGG
 
 | |
