4I3F
 
 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
6I8F
 
 | |
7ZR3
 
 | |
3V77
 
 | | Crystal structure of a putative fumarylacetoacetate isomerase/hydrolase from Oleispira antarctica | | Descriptor: | ACETATE ION, D(-)-TARTARIC ACID, Putative fumarylacetoacetate isomerase/hydrolase, ... | | Authors: | Stogios, P.J, Kagan, O, Di Leo, R, Bochkarev, A, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3VCR
 
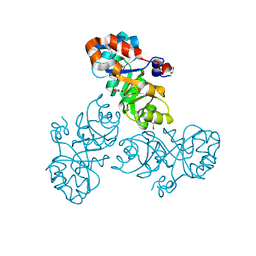 | | Crystal structure of a putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase from Oleispira antarctica | | Descriptor: | PYRUVIC ACID, putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase | | Authors: | Stogios, P.J, Kagan, O, Di Leo, R, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3QVM
 
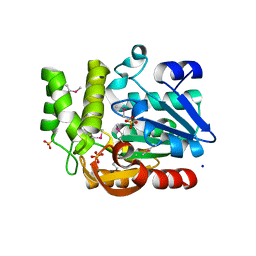 | | The structure of olei00960, a hydrolase from Oleispira antarctica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Olei00960, ... | | Authors: | Singer, A.U, Kagan, O, Kim, Y, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3S8Y
 
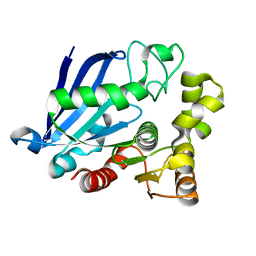 | | Bromide soaked structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | BROMIDE ION, Esterase APC40077 | | Authors: | Petit, P, Dong, A, Kagan, O, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
5K6N
 
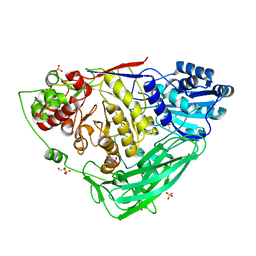 | |
5K6O
 
 | |
5K6M
 
 | |
5K6L
 
 | |
6RB0
 
 | |
6RKY
 
 | |
6SXY
 
 | |
6SYA
 
 | |
3I4Q
 
 | | Structure of a putative inorganic pyrophosphatase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | APC40078, SODIUM ION | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3I6Y
 
 | | Structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
3IRU
 
 | | Crystal structure of phoshonoacetaldehyde hydrolase like protein from Oleispira antarctica | | Descriptor: | SODIUM ION, phoshonoacetaldehyde hydrolase like protein | | Authors: | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3LMB
 
 | | The crystal structure of the protein OLEI01261 with unknown function from Chlorobaculum tepidum TLS | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, R, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | Descriptor: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | Authors: | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-02 | | Release date: | 2010-02-16 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | Descriptor: | Transaldolase | | Authors: | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
4Q3M
 
 | | Crystal structure of MGS-M4, an aldo-keto reductase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | MGS-M4, SODIUM ION, SULFATE ION | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3K
 
 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3N
 
 | | Crystal structure of MGS-M5, a lactate dehydrogenase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
