7SG4
 
 | |
7KDH
 
 | |
7KEB
 
 | |
7KDI
 
 | |
7KEA
 
 | |
7KDG
 
 | |
7KDL
 
 | |
7KE8
 
 | |
7KE9
 
 | |
7KE6
 
 | |
7KE4
 
 | |
7KDK
 
 | |
7KDJ
 
 | |
7KEC
 
 | |
7KE7
 
 | |
4QY6
 
 | | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations | | Descriptor: | Beta-lactamase TEM, Beta-lactamase PSE-4, CHLORIDE ION, ... | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations
To be Published
|
|
4QY5
 
 | | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations | | Descriptor: | Beta-lactamase TEM,Beta-lactamase PSE-4, CHLORIDE ION, MAGNESIUM ION | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations
To be Published
|
|
8CSA
 
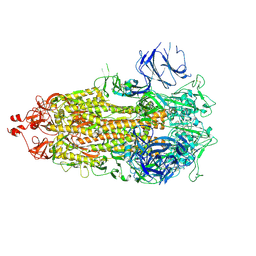 | |
8DKF
 
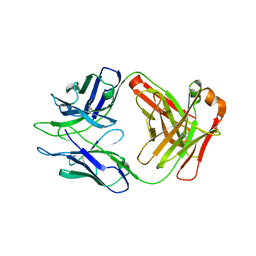 | | Antibody DH1030.1 Fab fragment | | Descriptor: | DH1030.1 Heavy chain, DH1030.1 Light chain | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DH1030.1 glycan reactive Ab
To Be Published
|
|
8DP1
 
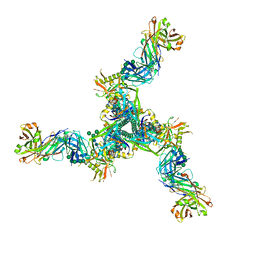 | | Cryo-EM structure of HIV-1 Env(BG505.T332N SOSIP) in complex with DH1030.1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1030.1 Fab Heavy chain, ... | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
To Be Published
|
|
8DOW
 
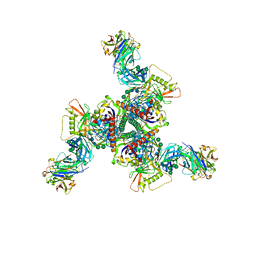 | | Cryo-EM structure of HIV-1 Env(CH848 10.17 DS.SOSIP_DT) in complex with DH1030.1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1030.1 Fab Heavy chain, ... | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
To Be Published
|
|
6VCU
 
 | | Homo sapiens FKBP12 protein bound with APX879 in P32 space group | | Descriptor: | ACETATE ION, N'-[(3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,26aS)-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-1,20,21-trioxo-8-(prop-2-en-1-yl)-1,3,4,5,6,8,11,12,13,14,15,16,17,18,19,20,21,23,24,25,26,26a-docosahydro-7H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosin-7-ylidene]acetohydrazide, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Gobeil, S, Spicer, L. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs.
Mbio, 12, 2021
|
|
6VCT
 
 | | Mucor circinelloides FKBP12 protein bound with APX879 in C2221 space group | | Descriptor: | N'-[(3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,26aS)-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-1,20,21-trioxo-8-(prop-2-en-1-yl)-1,3,4,5,6,8,11,12,13,14,15,16,17,18,19,20,21,23,24,25,26,26a-docosahydro-7H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosin-7-ylidene]acetohydrazide, Peptidylprolyl isomerase | | Authors: | Gobeil, S, Spicer, L. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs.
Mbio, 12, 2021
|
|
6VCV
 
 | | Aspergillus fumigatus FKBP12 protein bound with APX879 in P1 space group | | Descriptor: | FK506-binding protein 1A, N'-[(3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,26aS)-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-1,20,21-trioxo-8-(prop-2-en-1-yl)-1,3,4,5,6,8,11,12,13,14,15,16,17,18,19,20,21,23,24,25,26,26a-docosahydro-7H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosin-7-ylidene]acetohydrazide | | Authors: | Gobeil, S, Spicer, L. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs.
Mbio, 12, 2021
|
|
6VRX
 
 | |
