1OEB
 
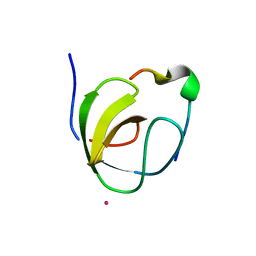 | | Mona/Gads SH3C domain | | Descriptor: | CADMIUM ION, GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Harkiolaki, M, Lewitzky, M, Gilbert, R.J.C, Jones, E.Y, Bourette, R.P, Mouchiroud, G, Sondermann, H, Moarefi, I, Feller, S.M. | | Deposit date: | 2003-03-24 | | Release date: | 2003-04-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for SH3 Domain-Mediated High-Affinity Binding between Mona/Gads and Slp-76
Embo J., 22, 2003
|
|
3OSK
 
 | | Crystal structure of human CTLA-4 apo homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Yu, C, Sonnen, A.F.-P, Ikemizu, S, Stuart, D.I, Gilbert, R.J.C, Davis, S.J. | | Deposit date: | 2010-09-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering
J.Biol.Chem., 286, 2011
|
|
4UD5
 
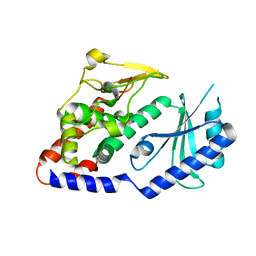 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Plasticity of Cid1 Provides a Basis for its Distributive RNA Terminal Uridylyl Transferase Activity.
Nucleic Acids Res., 43, 2015
|
|
8X7T
 
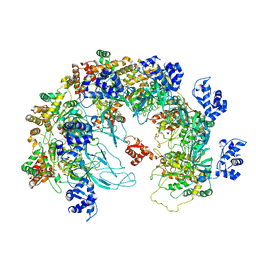 | | MCM in the Apo state. | | Descriptor: | mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | MCM in the Apo state
To Be Published
|
|
8X7U
 
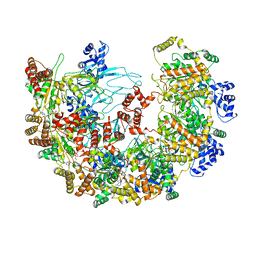 | | MCM in complex with dsDNA in presence of ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | MCM in complex with dsDNA in presence of ATP
To Be Published
|
|
6SB1
 
 | | Crystal structure of murine perforin-2 P2 domain crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Macrophage-expressed gene 1 protein | | Authors: | Ni, T, Ginger, L, Gilbert, R.J.C. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity.
Sci Adv, 6, 2020
|
|
6SB3
 
 | |
6SB4
 
 | | Crystal structure of murine perforin-2 P2 domain crystal form 2 | | Descriptor: | Macrophage-expressed gene 1 protein | | Authors: | Ni, T, Yu, X, Ginger, L, Gilbert, R.J.C. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity.
Sci Adv, 6, 2020
|
|
6SB5
 
 | |
1YJD
 
 | | Crystal structure of human CD28 in complex with the Fab fragment of a mitogenic antibody (5.11A1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment of 5.11A1 antibody heavy chain, Fab fragment of 5.11A1 antibody light chain, ... | | Authors: | Evans, E.J, Esnouf, R.M, Manso-Sancho, R, Gilbert, R.J.C, James, J.R, Sorensen, P, Stuart, D.I, Davis, S.J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a soluble CD28-Fab complex
Nat.Immunol., 6, 2005
|
|
7PGK
 
 | | HHIP-N, the N-terminal domain of the Hedgehog-Interacting Protein (HHIP), in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Hedgehog-interacting protein | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
7PGN
 
 | | HHP-C in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
7PGL
 
 | | HHIP-N, the N-terminal domain of the Hedgehog-Interacting Protein (HHIP), apo-form | | Descriptor: | Hedgehog-interacting protein | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
7PGM
 
 | | HHIP-C in complex with heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-3)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hedgehog-interacting protein | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
5OWN
 
 | | Structure of TgPLP1 MACPF domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
5OUP
 
 | | Structure of TgPLP1 MACPF domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
5OUO
 
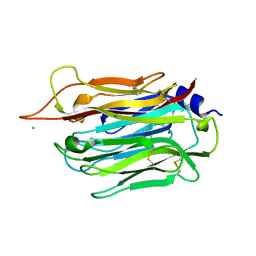 | | Structure of TgPLP1 APCbeta domain | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
5OUQ
 
 | | Structure of TgPLP1 MACPF domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.11 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
5OPG
 
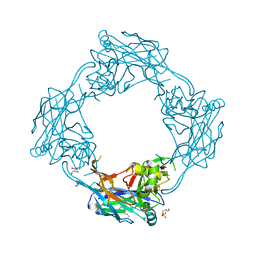 | | Structure of the Hantaan virus Gn glycoprotein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Rissanen, I, Stass, R, Zeltina, A, Li, S, Hepojoki, J, Harlos, K, Gilbert, R.J.C, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2017-08-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Transitions of the Conserved and Metastable Hantaviral Glycoprotein Envelope.
J. Virol., 91, 2017
|
|
2BPE
 
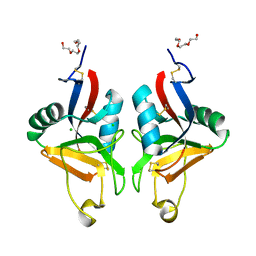 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECTIN-1, ... | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2013-01-16 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2017-04-19 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BPD
 
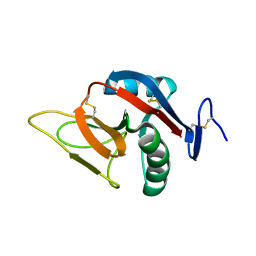 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | DECTIN-1 | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2BPH
 
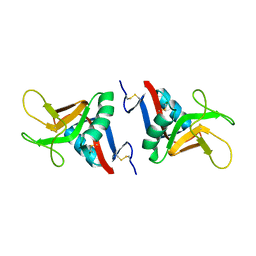 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | DECTIN-1, MAGNESIUM ION | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2CME
 
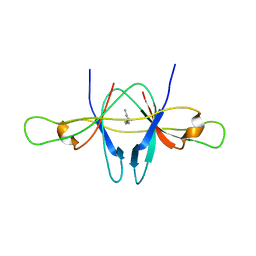 | | The crystal structure of SARS coronavirus ORF-9b protein | | Descriptor: | DECANE, HYPOTHETICAL PROTEIN 5 | | Authors: | Meier, C, Aricescu, A.R, Assenberg, R, Aplin, R.T, Gilbert, R.J.C, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2006-05-06 | | Release date: | 2006-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Orf-9B, a Lipid Binding Protein from the Sars Coronavirus.
Structure, 14, 2006
|
|
