4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
5KW2
 
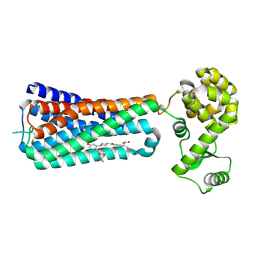 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
4MI5
 
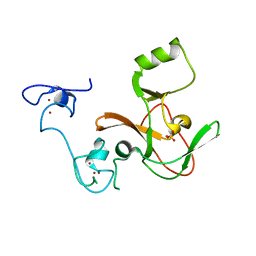 | | Crystal structure of the EZH2 SET domain | | Descriptor: | Histone-lysine N-methyltransferase EZH2, SULFATE ION, ZINC ION | | Authors: | Antonysamy, S, Condon, B, Druzina, Z, Bonanno, J, Gheyi, T, Macewan, I, Zhang, A, Ashok, S, Russell, M, Luz, J.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Context of Disease-Associated Mutations and Putative Mechanism of Autoinhibition Revealed by X-Ray Crystallographic Analysis of the EZH2-SET Domain.
Plos One, 8, 2013
|
|
4GQB
 
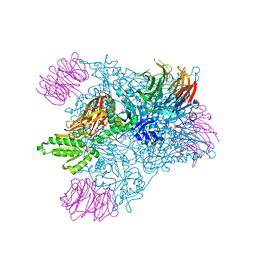 | | Crystal Structure of the human PRMT5:MEP50 Complex | | Descriptor: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | Authors: | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4I5I
 
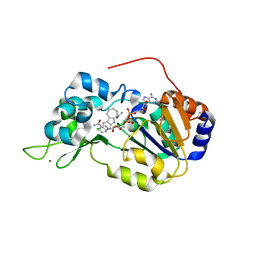 | | Crystal structure of the SIRT1 catalytic domain bound to NAD and an EX527 analog | | Descriptor: | (6S)-2-chloro-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhao, X, Allison, D, Condon, B, Zhang, F, Gheyi, T, Zhang, A, Ashok, S, Russell, M, Macewan, I, Qian, Y, Jamison, J.A, Luz, J.G. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.
J.Med.Chem., 56, 2013
|
|
3NF5
 
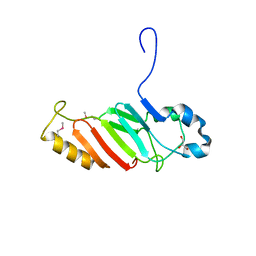 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
3BGE
 
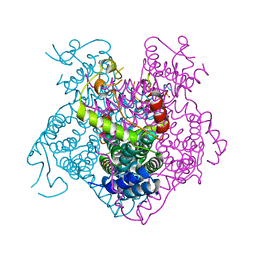 | | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae | | Descriptor: | Predicted ATPase, SULFATE ION | | Authors: | Ramagopal, U.A, Patskovsky, Y, Bonanno, J.B, Meyer, A.J, Toro, R, Freeman, J, Adams, J, Koss, J, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-26 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae.
To be Published
|
|
3BH1
 
 | | Crystal structure of protein DIP2346 from Corynebacterium diphtheriae | | Descriptor: | GLYCEROL, UPF0371 protein DIP2346 | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Gilmore, M, Iizuka, M, Groshong, C, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of protein DIP2346 from Corynebacterium diphtheriae.
To be Published
|
|
2GDQ
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution | | Descriptor: | yitF | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-16 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution
To be Published
|
|
2GGE
 
 | | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, yitF | | Authors: | Malashkevich, V.N, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A
To be Published
|
|
2IA1
 
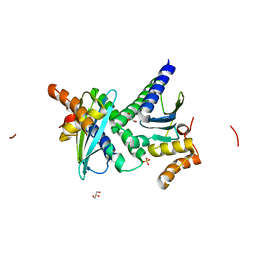 | | Crystal structure of protein BH3703 from Bacillus halodurans, Pfam DUF600 | | Descriptor: | BH3703 protein, GLYCEROL, SULFATE ION | | Authors: | Ramagopal, U.A, Russell, M, Toro, R, Freeman, J.C, Reyes, C, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of hypothetical protein BH3703 from Bacillus halodurans
To be Published
|
|
2IJQ
 
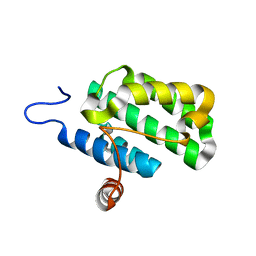 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
3H9M
 
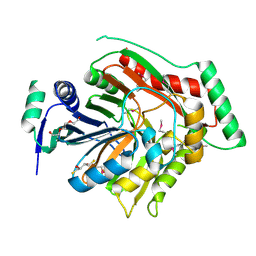 | | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, TRIETHYLENE GLYCOL, p-aminobenzoate synthetase, ... | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Do, J, Bain, K, Rutter, M, Gheyi, T, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii
To be Published
|
|
3HQC
 
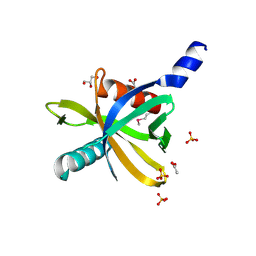 | | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sampathkumar, P, Romero, R, Wasserman, S, Do, J, Dickey, M, Bain, K, Gheyi, T, Klemke, R, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1)
To be Published
|
|
3HWJ
 
 | | Crystal structure of the second PHR domain of Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | DIMETHYL SULFOXIDE, E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Miller, S.A, Bain, K.T, Rutter, M.E, Gheyi, T, Klemke, R.L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
3JTY
 
 | | Crystal structure of a BenF-like porin from Pseudomonas fluorescens Pf-5 | | Descriptor: | BenF-like porin, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Sampathkumar, P, Lu, F, Zhao, X, Wasserman, S, Iuzuka, M, Bain, K, Rutter, M, Gheyi, T, Atwell, S, Luz, J, Gilmore, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution.
Proteins, 78, 2010
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3BJS
 
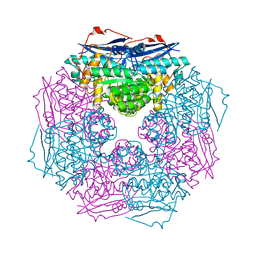 | | Crystal structure of a member of enolase superfamily from Polaromonas sp. JS666 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Patskovsky, Y, Bonanno, J.B, Ozyurt, S, Dickey, M, Sauder, J.M, Reyes, C, Groshong, C, Gheyi, T, Smith, D, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Member of Enolase Superfamily from Polaromonas sp. JS666.
To be Published
|
|
3BMA
 
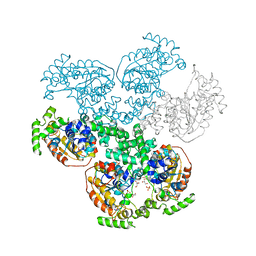 | | Crystal structure of D-alanyl-lipoteichoic acid synthetase from Streptococcus pneumoniae R6 | | Descriptor: | D-alanyl-lipoteichoic acid synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Smith, D, Rutter, M, Iizuka, M, Koss, J, Bain, K, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of probable D-Alanyl-Lipoteichoic Acid Synthetase from Streptococcus pneumoniae.
To be Published
|
|
3BT5
 
 | | Crystal structure of DUF305 fragment from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, Uncharacterized protein DUF305 | | Authors: | Ramagopal, U.A, Patskovsky, Y, Rutter, M, Toro, R, Bain, K, Meyer, A.J, Powell, A, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-27 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of DUF305 fragment from Deinococcus radiodurans.
To be Published
|
|
3CMG
 
 | | Crystal structure of putative beta-galactosidase from Bacteroides fragilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Ramagopal, U.A, Rutter, M, Toro, R, Hu, S, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-21 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative beta-galactosidase from Bacteroides fragilis.
To be published
|
|
3CZB
 
 | | Crystal structure of putative transglycosylase from Caulobacter crescentus | | Descriptor: | Putative transglycosylase, SULFATE ION | | Authors: | Ramagopal, U.A, Chattopadhyay, K, Toro, R, Wasserman, S, Freeman, J, Logan, C, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-06-10 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative transglycosylase from Caulobacter crescentus.
To be Published
|
|
3DLS
 
 | | Crystal structure of human PAS kinase bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PAS domain-containing serine/threonine-protein kinase | | Authors: | Antonysamy, S, Bonanno, J.B, Romero, R, Russell, M, Iizuka, M, Gheyi, T, Wasserman, S.R, Rutter, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-29 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural bases of PAS domain-regulated kinase (PASK) activation in the absence of activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
