6DCM
 
 | |
8E8Q
 
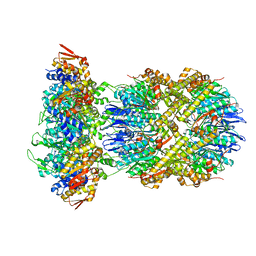 | | Cryo-EM structure of substrate-free DNClpX.ClpP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8E7V
 
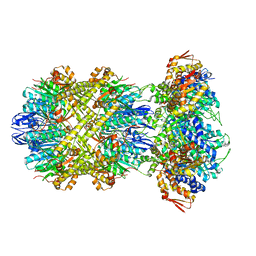 | | Cryo-EM structure of substrate-free DNClpX.ClpP from singly capped particles | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8E91
 
 | |
8ET3
 
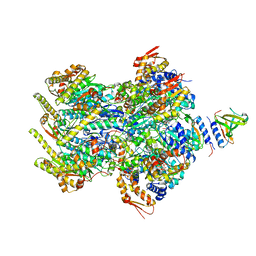 | | Cryo-EM structure of a delivery complex containing the SspB adaptor, an ssrA-tagged substrate, and the AAA+ ClpXP protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Ghanbarpour, A, Fei, X, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-10-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6WP0
 
 | |
6MQW
 
 | |
6NOE
 
 | |
6NNX
 
 | |
6NNY
 
 | |
6ON5
 
 | |
6ON8
 
 | |
6ON7
 
 | |
7UCN
 
 | |
7UCS
 
 | |
7UCT
 
 | |
7UCV
 
 | |
7UCZ
 
 | |
7UD1
 
 | |
7UD3
 
 | |
6VIT
 
 | |
6VID
 
 | |
6VIS
 
 | |
6WNF
 
 | |
6WNJ
 
 | |
