4MOD
 
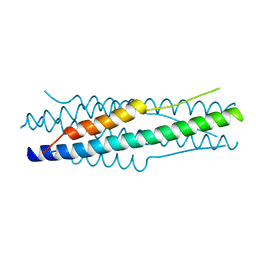 | | Structure of the MERS-CoV fusion core | | Descriptor: | HR1 of S protein, LINKER, HR2 of S protein | | Authors: | Gao, J, Lu, G, Qi, J, Li, Y, Wu, Y, Deng, Y, Geng, H, Xiao, H, Tan, W, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
6XRT
 
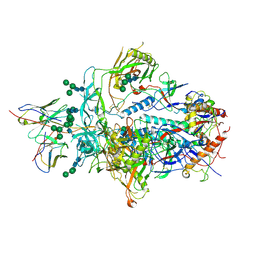 | |
6DE7
 
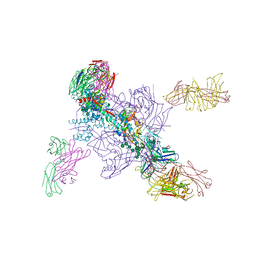 | |
6MQE
 
 | |
6MQS
 
 | |
6NF2
 
 | |
6OSY
 
 | |
6OT1
 
 | |
6CDO
 
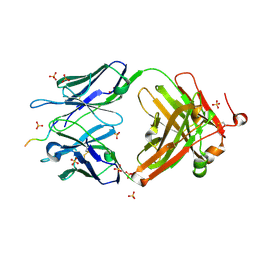 | | Structure of vaccine-elicited HIV-1 neutralizing antibody vFP16.02 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP16.02 Fab heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDP
 
 | | Vaccine-elicited HIV-1 neutralizing antibody vFP20.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP20.01 Fab heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6XCJ
 
 | | Crystal Structure of DH650 Fab from a Rhesus Macaque in Complex with HIV-1 gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH650 Fab Heavy Chain, DH650 Fab Light Chain, ... | | Authors: | Raymond, D.D, Chug, H, Harrison, S.C. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth.
Science, 371, 2021
|
|
6VIL
 
 | |
5TKJ
 
 | | Structure of vaccine-elicited diverse HIV-1 neutralizing antibody vFP1.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, vFP1.01 chimeric mouse antibody heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2016-10-06 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
5TKK
 
 | |
8WD6
 
 | | Cryo-EM structure of the ABCG25 | | Descriptor: | ABC transporter G family member 25 | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
8WAM
 
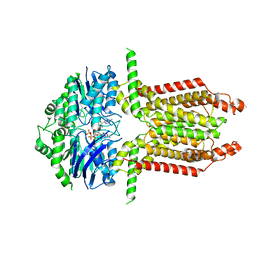 | |
8WBA
 
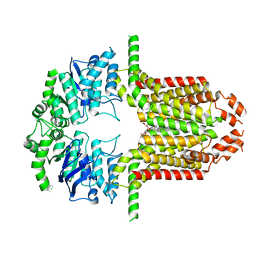 | | Cryo-EM structure of the ABCG25 bound to CHS | | Descriptor: | ABC transporter G family member 25, CHOLESTEROL HEMISUCCINATE | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-09 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
8WBX
 
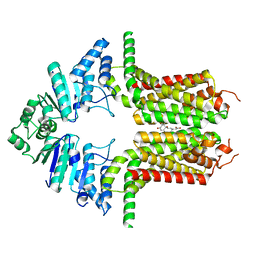 | | Cryo-EM structure of the ABCG25 bound to ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-10 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
6VRW
 
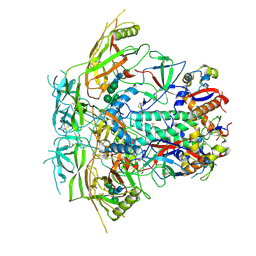 | | Cryo-EM structure of stabilized HIV-1 Env trimer CAP256.wk34.c80 SOSIP.RnS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-03-11 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structure of Super-Potent Antibody CAP256-VRC26.25 in Complex with HIV-1 Envelope Reveals a Combined Mode of Trimer-Apex Recognition.
Cell Rep, 31, 2020
|
|
6VZI
 
 | |
6VTT
 
 | |
6W03
 
 | |
6WWC
 
 | |
6WX2
 
 | |
5U7O
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-626529 in Complex with Human Antibodies PGT122 and 35O22 at 3.8 Angstrom | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pancera, M, Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.031 Å) | | Cite: | Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529.
Nat. Chem. Biol., 13, 2017
|
|
