225D
 
 | |
8CT8
 
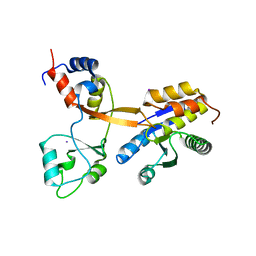 | | Crystal structure of Drosophila melanogaster PRL/CBS-pair domain complex | | Descriptor: | IODIDE ION, PRL-1 phosphatase, Unextended protein | | Authors: | Fakih, R, Goldstein, R.H, Kozlov, G, Gehring, K. | | Deposit date: | 2022-05-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Burst kinetics and CNNM binding are evolutionarily conserved properties of phosphatases of regenerating liver.
J.Biol.Chem., 299, 2023
|
|
7SOW
 
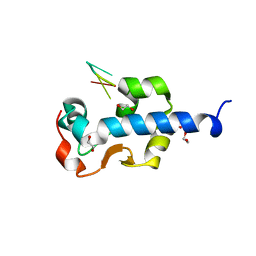 | | LaM domain of human LARP1 in complex with UUUUUU | | Descriptor: | 1,2-ETHANEDIOL, Isoform 2 of La-related protein 1, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of 3'-end RNA recognition by LARP1
To Be Published
|
|
7US1
 
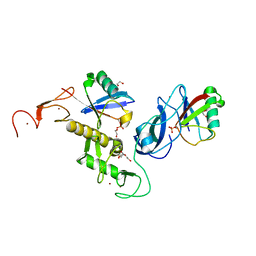 | | Structure of parkin (R0RB) bound to two phospho-ubiquitin molecules | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin, ... | | Authors: | Fakih, R, Sauve, V, Gehring, K. | | Deposit date: | 2022-04-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Structure of the second phosphoubiquitin-binding site in parkin.
J.Biol.Chem., 298, 2022
|
|
8GK6
 
 | |
4K7D
 
 | | Crystal Structure of Parkin C-terminal RING domains | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, MALONATE ION, ... | | Authors: | Sauve, V, Trempe, J.-F, Menade, M, Gehring, K. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
4K95
 
 | | Crystal Structure of Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Seirafi, M, Menade, M, Sauve, V, Kozlov, G, Trempe, J.-F, Nagar, B, Gehring, K. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
6B3Y
 
 | | Crystal structure of the PH-like domain from DENND3 | | Descriptor: | DENN domain-containing protein 3 | | Authors: | Kozlov, G, Xu, J, Menade, M, Beaugrand, M, Pan, T, McPherson, P.S, Gehring, K. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | A PH-like domain of the Rab12 guanine nucleotide exchange factor DENND3 binds actin and is required for autophagy.
J. Biol. Chem., 293, 2018
|
|
7MSU
 
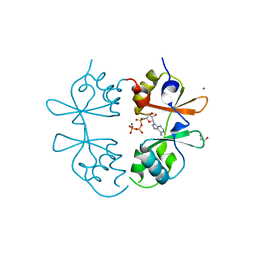 | | Crystal structure of an archaeal CNNM, MtCorB, CBS-pair domain in complex with Mg2+-ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Hemolysin, ... | | Authors: | Chen, Y.S, Gehring, K. | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of an archaeal CorB magnesium transporter.
Nat Commun, 12, 2021
|
|
6MN6
 
 | |
5KDG
 
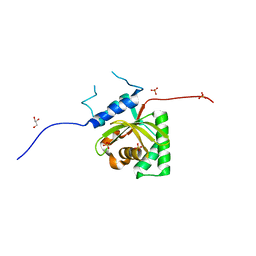 | | Crystal Structure of Salmonella Typhimurium Effector GtgE | | Descriptor: | GLYCEROL, Gifsy-2 prophage protein, SULFATE ION | | Authors: | Kozlov, G, Xu, C, Wong, K, Gehring, K, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the Salmonella Typhimurium Effector GtgE.
PLoS ONE, 11, 2016
|
|
4Z2Z
 
 | |
4ZYN
 
 | | Crystal Structure of Parkin E3 ubiquitin ligase (linker deletion; delta 86-130) | | Descriptor: | E3 ubiquitin-protein ligase parkin, SULFATE ION, ZINC ION | | Authors: | Lilov, A, Sauve, V, Trempe, J.F, Rodionov, D, Wang, J, Gehring, K. | | Deposit date: | 2015-05-21 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A Ubl/ubiquitin switch in the activation of Parkin.
Embo J., 34, 2015
|
|
5K35
 
 | | Structure of the Legionella effector, AnkB, in complex with human Skp1 | | Descriptor: | Ankyrin-repeat protein B, S-phase kinase-associated protein 1 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5K34
 
 | | Structure of the ankyrin domain of AnkB from Legionella Pneumophila | | Descriptor: | Ankyrin-repeat protein B, GLYCEROL, SULFATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5KES
 
 | | Solution structure of the yeast Ddi1 HDD domain | | Descriptor: | DNA damage-inducible protein 1 | | Authors: | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | Deposit date: | 2016-06-10 | | Release date: | 2016-10-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
8F6D
 
 | |
8EY7
 
 | |
8EY6
 
 | |
8EY8
 
 | |
8G91
 
 | |
8G90
 
 | |
6WUS
 
 | |
6WUR
 
 | |
6PIK
 
 | | Tetrameric cryo-EM ArnA | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
