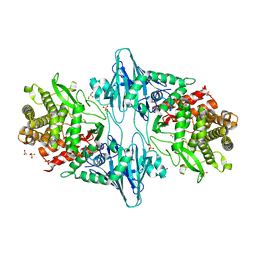
8QME
 
 | |
2OLP
 
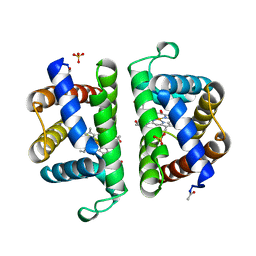 | | Structure and ligand selection of hemoglobin II from Lucina pectinata | | Descriptor: | Hemoglobin II, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gavira, J.A, Camara-Artigas, A, de Jesus, W, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2007-01-19 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structure and Ligand Selection of Hemoglobin II from Lucina pectinata
J.Biol.Chem., 283, 2008
|
|
5T65
 
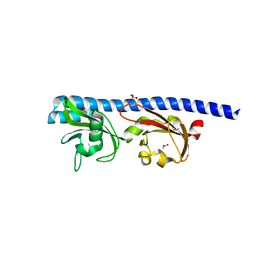 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-ILE | | Descriptor: | ACETATE ION, ISOLEUCINE, Methyl-accepting chemotaxis protein PctA, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-01 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5T7M
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-TRP | | Descriptor: | ACETATE ION, Chemotaxis protein, SODIUM ION, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LT9
 
 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Arg | | Descriptor: | ARGININE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LTO
 
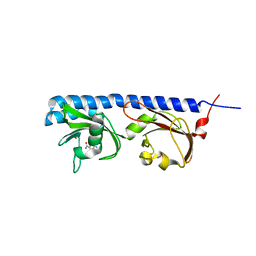 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Gln | | Descriptor: | GLUTAMINE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.459 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
2BDZ
 
 | | Mexicain from Jacaratia mexicana | | Descriptor: | Mexicain, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Gavira, J.A, Oliver-Salvador, M.C, Gonzalez-Ramirez, L.A, Soriano-Garcia, M, Garcia-Ruiz, J.M. | | Deposit date: | 2005-10-21 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic structure of Mexicain from Jacaratia mexicana
To be Published
|
|
5LTX
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-MET | | Descriptor: | ACETATE ION, Chemotaxis protein, FORMIC ACID, ... | | Authors: | Gavira, J.A, Rico-Gimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LTV
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECPETOR PCTC IN COMPLEX WITH GABA | | Descriptor: | ACETATE ION, Chemotactic transducer PctC, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
1Z2U
 
 | | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gavira, J.A, DiGiamamarino, E, Tempel, W, Liu, Z.J, Wang, B.C, Meehan, E, Ng, J.D, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance
To be published
|
|
1Z3D
 
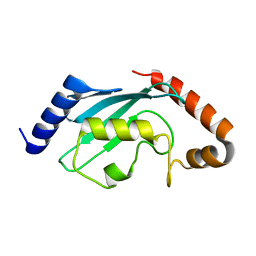 | | Protein crystal growth improvement leading to the 2.5A crystallographic structure of ubiquitin-conjugating enzyme (ubc-1) from Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2 1 | | Authors: | Gavira, J.A, DiGiammarino, E, Tempel, W, Toh, D, Liu, Z.J, Wang, B.C, Meehan, E, Ng, J.D, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-03-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein crystal growth improvement leading to the 2.5A crystallographic structure of ubiquitin-conjugating enzyme (ubc-1) from Caenorhabditis elegans
To be Published
|
|
1ZZY
 
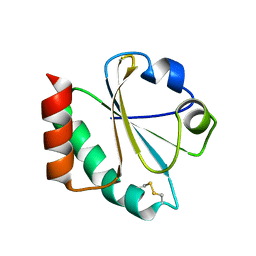 | |
6YC5
 
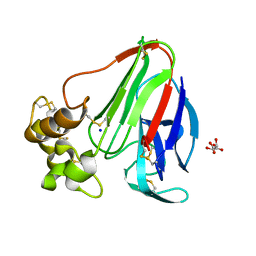 | |
6YBI
 
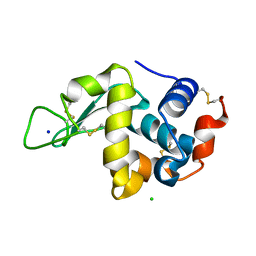 | |
6YBO
 
 | |
6YBX
 
 | |
6YBR
 
 | |
6S33
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with Protocatechuate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S38
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S1A
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP | | Descriptor: | Aromatic acid chemoreceptor, SULFATE ION | | Authors: | Gavira, J.A, Matilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S37
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, ACETATE ION, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S18
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with glycerol | | Descriptor: | Aromatic acid chemoreceptor, CHLORIDE ION, GLYCEROL | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-10-21 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S3B
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with benzoate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aromatic acid chemoreceptor, ... | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
7PTJ
 
 | |
7PTH
 
 | |
