4ACI
 
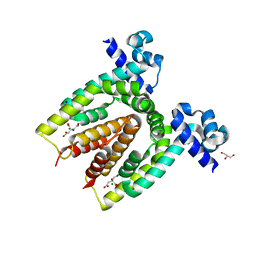 | | Structure of the C. glutamicum AcnR Crystal Form II | | Descriptor: | CITRIC ACID, GLYCEROL, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR, ... | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
4AF5
 
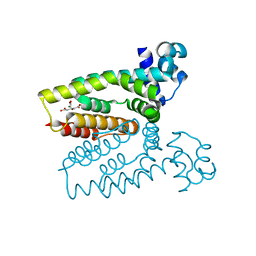 | | Structure of the C. glutamicum AcnR Crystal Form I | | Descriptor: | CITRIC ACID, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR COMPND 2, MAGNESIUM ION | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
4AOO
 
 | | B. subtilis dUTPase YncF in complex with dU PPi and Mg in H32 | | Descriptor: | 2'-DEOXYURIDINE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Garcia-Nafria, J, Timm, J, Harrison, C, Turkenburg, J.P, Wilson, K.S. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tying Down the Arm in Bacillus Dutpase: Structure and Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4APZ
 
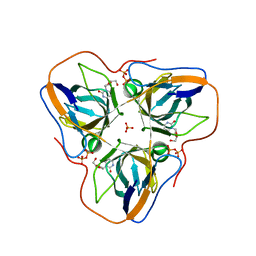 | | Structure of B. subtilis genomic dUTPase YncF in complex with dU, PPi and Mg in P1 | | Descriptor: | 2'-DEOXYURIDINE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Garcia-Nafria, J, Timm, J, Harrison, C, Turkenburg, J.P, Wilson, K.S. | | Deposit date: | 2012-04-11 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Tying Down the Arm in Bacillus Dutpase: Structure and Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2Y1T
 
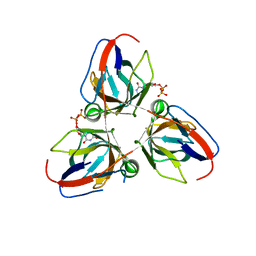 | | Bacillus subtilis prophage dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XCE
 
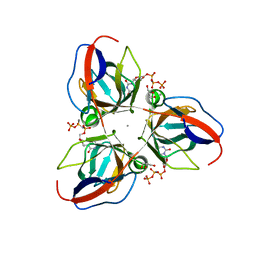 | | Structure of YncF in complex with dUpNHpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Garcia-Nafria, J, Burchell, L, Takezawa, M, Rzechorzek, N, Fogg, M, Wilson, K.S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Genomic Bacillus Subtilis Dutpase: Novel Features in the Phe-Lid.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2XX6
 
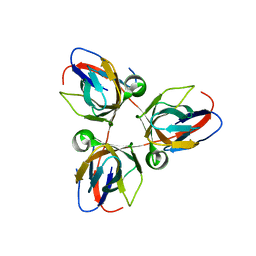 | | Structure of the Bacillus subtilis prophage dUTPase, YosS | | Descriptor: | SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-11-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XY3
 
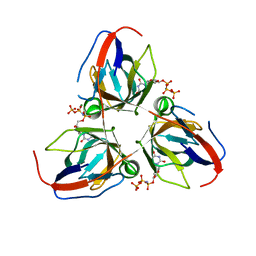 | | Structure of the Bacillus subtilis prophage dUTPase with dUpNHpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, MAGNESIUM ION, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4AO5
 
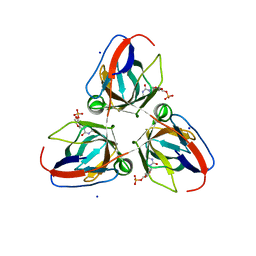 | | B. subtilis prophage dUTPase YosS in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, SODIUM ION, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEO TIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harrison, C, Turkenburg, J.P, Wilson, K.S. | | Deposit date: | 2012-03-23 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tying Down the Arm in Bacillus Dutpase: Structure and Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6G79
 
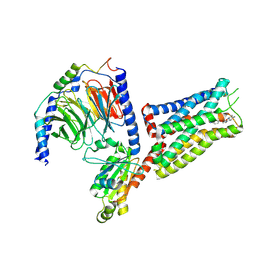 | | Coupling specificity of heterotrimeric Go to the serotonin 5-HT1B receptor | | Descriptor: | 2-[5-[2-[4-(4-cyanophenyl)piperazin-1-yl]-2-oxidanylidene-ethoxy]-1~{H}-indol-3-yl]ethylazanium, 5-hydroxytryptamine receptor 1B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Garcia-Nafria, J, Nehme, R, Edwards, P, Tate, C.G. | | Deposit date: | 2018-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM structure of the serotonin 5-HT1Breceptor coupled to heterotrimeric Go.
Nature, 558, 2018
|
|
6GDG
 
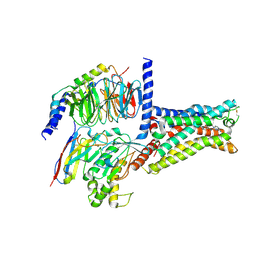 | | Cryo-EM structure of the adenosine A2A receptor bound to a miniGs heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Garcia-Nafria, J, Lee, Y. | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM structure of the adenosine A2Areceptor coupled to an engineered heterotrimeric G protein.
Elife, 7, 2018
|
|
4AC6
 
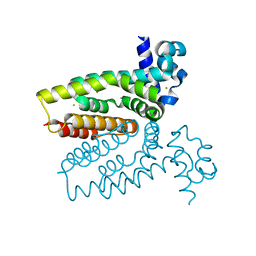 | | Corynebacterium glutamicum AcnR AU derivative structure | | Descriptor: | GOLD ION, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-26 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
5FWX
 
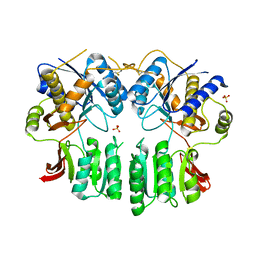 | | Crystal structure of the AMPA receptor GluA2/A4 N-terminal domain heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMATE RECEPTOR 2, GLUTAMATE RECEPTOR 4, ... | | Authors: | Garcia-Nafria, J, Herguedas, B, Greger, I.H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Organization of Heteromeric Ampa-Type Glutamate Receptors.
Science, 352, 2016
|
|
6FLR
 
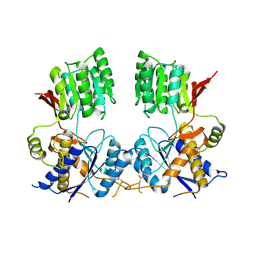 | | Super-open structure of the AMPAR GluA3 N-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 3 | | Authors: | Garcia-Nafria, J. | | Deposit date: | 2018-01-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
4AOZ
 
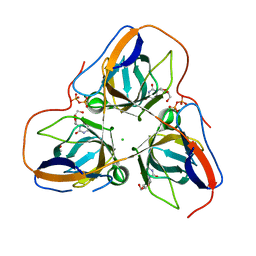 | | B. subtilis dUTPase YncF in complex with dU, PPi and Mg (P212121) | | Descriptor: | 2'-DEOXYURIDINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Timm, J, Garcia-Nafria, J, Harrison, C, Turkenburg, J.P, Wilson, K.S. | | Deposit date: | 2012-03-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tying Down the Arm in Bacillus Dutpase: Structure and Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4B0H
 
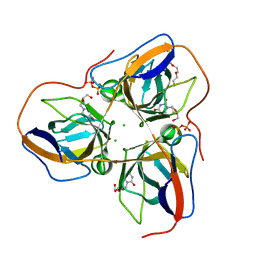 | | B. subtilis dUTPase YncF in complex with dU, PPi and Mg b (P212121) | | Descriptor: | 2'-DEOXYURIDINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Timm, J, Garcia-Nafria, J, Harrison, C, Turkenburg, J.P, Wilson, K.S. | | Deposit date: | 2012-07-02 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Tying Down the Arm in Bacillus Dutpase: Structure and Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6NIY
 
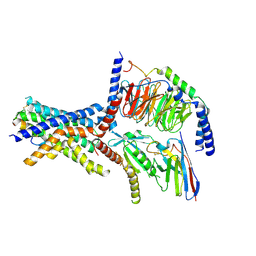 | | A high-resolution cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | dal Maso, E, Glukhova, A, Zhu, Y, Garcia-Nafria, J, Tate, C.G, Atanasio, S, Reynolds, C.A, Ramirez-Aportela, E, Carazo, J.-M, Hick, C.A, Furness, S.G.B, Hay, D.L, Liang, Y.-L, Miller, L.J, Christopoulos, A, Wang, M.-W, Wootten, D, Sexton, P.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-01-23 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | The Molecular Control of Calcitonin Receptor Signaling.
Acs Pharmacol Transl Sci, 2, 2019
|
|
5IDE
 
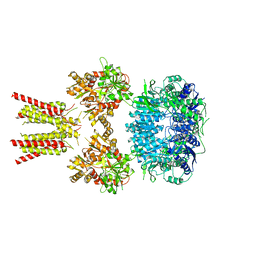 | | Cryo-EM structure of GluA2/3 AMPA receptor heterotetramer (model I) | | Descriptor: | Glutamate receptor 2, Glutamate receptor 3 | | Authors: | Herguedas, B, Garcia-Nafria, J, Fernandez-Leiro, R, Greger, I.H. | | Deposit date: | 2016-02-24 | | Release date: | 2016-03-16 | | Last modified: | 2017-10-11 | | Method: | ELECTRON MICROSCOPY (8.25 Å) | | Cite: | Structure and organization of heteromeric AMPA-type glutamate receptors.
Science, 352, 2016
|
|
5IDF
 
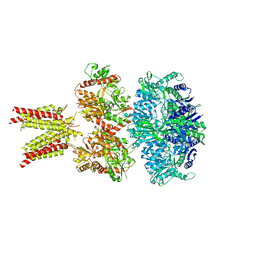 | | Cryo-EM structure of GluA2/3 AMPA receptor heterotetramer (model II) | | Descriptor: | Glutamate receptor 2, Glutamate receptor 3 | | Authors: | Herguedas, B, Garcia-Nafria, J, Fernandez-Leiro, R, Greger, I.H. | | Deposit date: | 2016-02-24 | | Release date: | 2016-03-16 | | Last modified: | 2017-10-11 | | Method: | ELECTRON MICROSCOPY (10.31 Å) | | Cite: | Structure and organization of heteromeric AMPA-type glutamate receptors.
Science, 352, 2016
|
|
5FWY
 
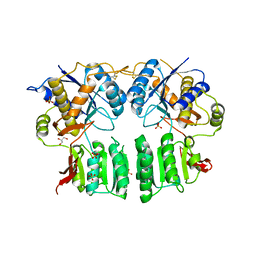 | | Crystal structure of the AMPA receptor GluA2/A3 N-terminal domain heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMATE RECEPTOR 2, GLUTAMATE RECEPTOR 3, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I.H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and Organization of Heteromeric Ampa-Type Glutamate Receptors.
Science, 352, 2016
|
|
6QKC
 
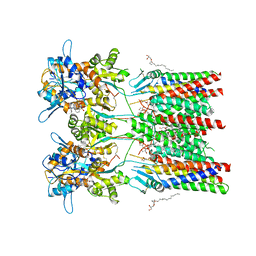 | | GluA1/2 In complex with auxiliary subunit gamma-8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 1, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I.G. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma 8.
Science, 364, 2019
|
|
6QKZ
 
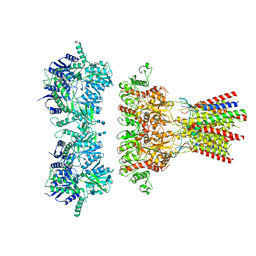 | | Full length GluA1/2-gamma8 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I.G. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma 8.
Science, 364, 2019
|
|
6FPJ
 
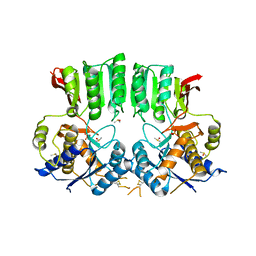 | | Structure of the AMPAR GluA3 N-terminal domain bound to phosphate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
6GBU
 
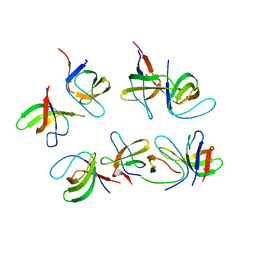 | | Crystal structure of the second SH3 domain of FCHSD2 (SH3-2) in complex with the fourth SH3 domain of ITSN1 (SH3d) | | Descriptor: | F-BAR and double SH3 domains protein 2, Intersectin-1 | | Authors: | Almeida-Souza, L, Frank, R, Garcia-Nafria, J, Colussi, A, Gunawardana, N, Johnson, C.M, Yu, M, Howard, G, Andrews, B, Vallis, Y, McMahon, H.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | A Flat BAR Protein Promotes Actin Polymerization at the Base of Clathrin-Coated Pits.
Cell, 174, 2018
|
|
2X36
 
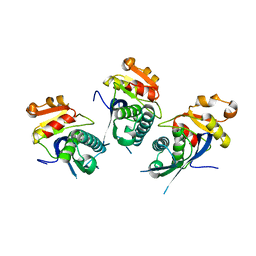 | | Structure of the proteolytic domain of the Human Mitochondrial Lon protease | | Descriptor: | LON PROTEASE HOMOLOG, MITOCHONDRIAL | | Authors: | Garcia, J, Ondrovicova, G, Blagova, E, Levdikov, V.M, Bauer, J.A, Kutejova, E, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2010-01-21 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Catalytic Domain of the Human Mitochondrial Lon Protease: Proposed Relation of Oligomer Formation and Activity.
Protein Sci., 19, 2010
|
|
