2WYL
 
 | | Apo structure of a metallo-b-lactamase | | Descriptor: | FORMYL GROUP, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
2WYM
 
 | | Structure of a metallo-b-lactamase | | Descriptor: | CITRATE ANION, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG, ... | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
4R26
 
 | | Crystal structure of human Fab PGT124, a broadly neutralizing and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGR124-Light Chain, PGT124-Heavy Chain | | Authors: | Garces, F, Kong, L, Wilson, I.A. | | Deposit date: | 2014-08-08 | | Release date: | 2014-10-08 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.4969 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
4R2G
 
 | | Crystal Structure of PGT124 Fab bound to HIV-1 JRCSF gp120 core and to CD4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Garces, F, Wilson, I.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
3ZVM
 
 | | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*GP*TP*CP*AP*CP)-3', ACETATE ION, ... | | Authors: | Garces, F, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase.
Mol. Cell, 44, 2011
|
|
3ZVN
 
 | | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase | | Descriptor: | 5'-D(*GP*TP*CP*AP*CP)-3', ADENOSINE-5'-DIPHOSPHATE, BIFUNCTIONAL POLYNUCLEOTIDE PHOSPHATASE/KINASE, ... | | Authors: | Garces, F, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase.
Mol. Cell, 44, 2011
|
|
3ZVL
 
 | | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase | | Descriptor: | ACETATE ION, BIFUNCTIONAL POLYNUCLEOTIDE PHOSPHATASE/KINASE, CHLORIDE ION, ... | | Authors: | Garces, F, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Structural Basis for Substrate Recognition by Mammalian Polynucleotide Kinase 3' Phosphatase.
Mol.Cell, 44, 2011
|
|
5CEX
 
 | |
5UM8
 
 | | Crystal structure of HIV-1 envelope trimer 16055 NFL TD CC (T569G) in complex with Fabs 35022 and PGT124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 35022 heavy chain, ... | | Authors: | Garces, F, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.935 Å) | | Cite: | Glycine Substitution at Helix-to-Coil Transitions Facilitates the Structural Determination of a Stabilized Subtype C HIV Envelope Glycoprotein.
Immunity, 46, 2017
|
|
5W6D
 
 | | Crystal structure of BG505-SOSIP.v4.1-GT1-N137A in complex with Fabs 35022 and 9H/109L | | Descriptor: | 109L FAB light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garces, F, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo.
J. Exp. Med., 214, 2017
|
|
5BZD
 
 | |
5BZW
 
 | |
5CEZ
 
 | |
5CEY
 
 | |
5NQ6
 
 | | Crystal structure of the inhibited form of the redox-sensitive SufE-like sulfur acceptor CsdE from Escherichia coli at 2.40 Angstrom Resolution | | Descriptor: | GLYCEROL, SULFATE ION, Sulfur acceptor protein CsdE | | Authors: | Penya-Soler, E, Aranda, J, Lopez-Estepa, M, Gomez, S, Garces, F, Coll, M, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the inhibited form of the redox-sensitive SufE-like sulfur acceptor CsdE.
PLoS ONE, 12, 2017
|
|
4CVQ
 
 | | CRYSTAL STRUCTURE OF AN AMINOTRANSFERASE FROM ESCHERICHIA COLI AT 2. 11 ANGSTROEM RESOLUTION | | Descriptor: | ACETATE ION, GLUTAMATE-PYRUVATE AMINOTRANSFERASE ALAA, GLYCEROL, ... | | Authors: | Penya-Soler, E, Fernandez, F.J, Lopez-Estepa, M, Garces, F, Richardson, A.J, Rudd, K.E, Coll, M, Vega, M.C. | | Deposit date: | 2014-03-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural analysis and mutant growth properties reveal distinctive enzymatic and cellular roles for the three major L-alanine transaminases of Escherichia coli.
PLoS ONE, 9, 2014
|
|
7LUS
 
 | | IgG2 Fc Charge Pair Mutation version 1 (CPMv1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 2 | | Authors: | Sudom, A, Whittington, D, Garces, F, Wang, Z. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Next generation Fc scaffold for multispecific antibodies.
Iscience, 24, 2021
|
|
7LUR
 
 | |
5FT4
 
 | | Crystal structure of the cysteine desulfurase CsdA from Escherichia coli at 1.996 Angstroem resolution | | Descriptor: | CITRIC ACID, CYSTEINE DESULFURASE CSDA, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Quintana, J.F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-12-21 | | Last modified: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | The Mechanism of Sulfur Transfer Across Protein- Protein Interfaces: The Csd Model
Acs Catalysis, 6, 2016
|
|
5FT8
 
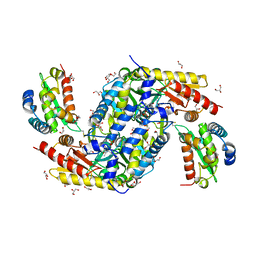 | | Crystal structure of the complex between the cysteine desulfurase CsdA and the sulfur-acceptor CsdE in the persulfurated state at 2.50 Angstroem resolution | | Descriptor: | Cysteine desulfurase CsdA, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
5FT5
 
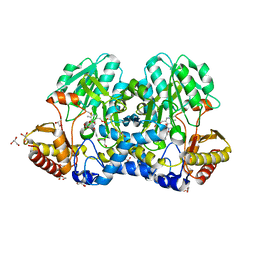 | | Crystal structure of the cysteine desulfurase CsdA (persulfurated) from Escherichia coli at 2.384 Angstroem resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Quintana, J.F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
5FT6
 
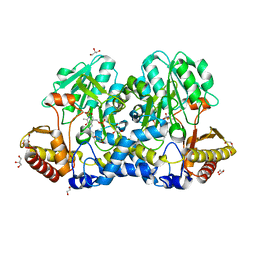 | | Crystal structure of the cysteine desulfurase CsdA (S-sulfonic acid) from Escherichia coli at 2.050 Angstroem resolution | | Descriptor: | CYSTEINE DESULFURASE CSDA, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
6UMI
 
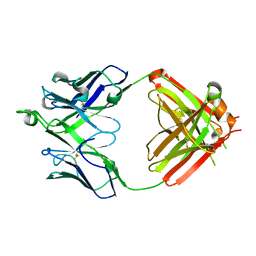 | | Crystal structure of erenumab Fab-b | | Descriptor: | erenumab Fab heavy chain, IgG1, erenumab Fab light chain | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMH
 
 | | Crystal structure of erenumab Fab-a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-PROPANDIOL, PHOSPHATE ION, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMJ
 
 | | Crystal structure of erenumab Fab-c | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-BUTANEDIOL, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
