1QVX
 
 | | SOLUTION STRUCTURE OF THE FAT DOMAIN OF FOCAL ADHESION KINASE | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Gao, G, Prutzman, K.C, King, M.L, DeRose, E.F, London, R.E, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Focal Adhesion Targeting Domain of Focal Adhesion Kinase in Complex with a Paxillin LD Peptide: EVIDENCE FOR A TWO-SITE BINDING MODEL.
J.Biol.Chem., 279, 2004
|
|
6U3Q
 
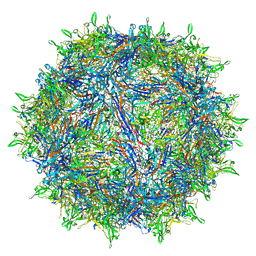 | | The atomic structure of a human adeno-associated virus capsid isolate (AAVhu69/AAVv66) | | Descriptor: | Capsid protein VP1 | | Authors: | Hsu, H.-L, Brown, A, Loveland, A, Tai, P, Korostelev, A, Gao, G. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties.
Nat Commun, 11, 2020
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
1PV3
 
 | | NMR Solution Structure of the Avian FAT-domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Prutzman, K.C, Gao, G, King, M.L, Iyer, V.V, Mueller, G.A, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-06-26 | | Release date: | 2004-05-25 | | Last modified: | 2016-04-27 | | Method: | SOLUTION NMR | | Cite: | The Focal Adhesion Targeting Domain of Focal Adhesion Kinase Contains a Hinge Region that Modulates Tyrosine 926 Phosphorylation.
STRUCTURE, 12, 2004
|
|
3U9G
 
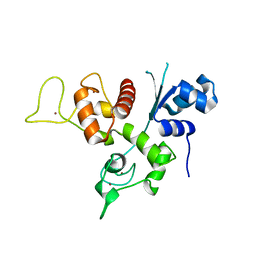 | | Crystal structure of the Zinc finger antiviral protein | | Descriptor: | ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Chen, S, Xu, Y, Zhang, K, Wang, X, Sun, J, Gao, G, Liu, Y. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V32
 
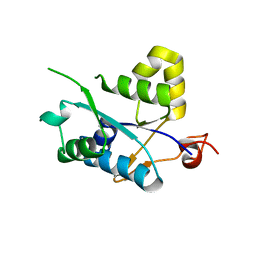 | | Crystal structure of MCPIP1 N-terminal conserved domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
3V33
 
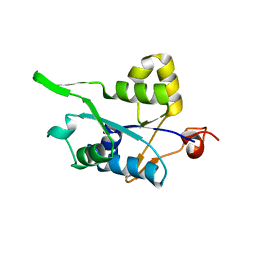 | | Crystal structure of MCPIP1 conserved domain with zinc-finger motif | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
3V34
 
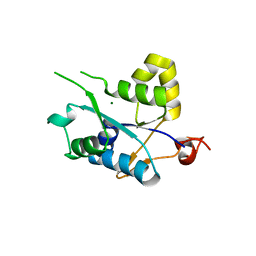 | | Crystal structure of MCPIP1 conserved domain with magnesium ion in the catalytic center | | Descriptor: | MAGNESIUM ION, Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
1TXD
 
 | |
6L1W
 
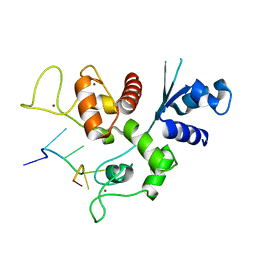 | | Zinc-finger Antiviral Protein (ZAP) bound to RNA | | Descriptor: | RNA (5'-R(*CP*GP*UP*CP*GP*U)-3'), ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Luo, X, Wang, X, Gao, Y, Zhu, J, Liu, S, Gao, G, Gao, P. | | Deposit date: | 2019-09-30 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Molecular Mechanism of RNA Recognition by Zinc-Finger Antiviral Protein.
Cell Rep, 30, 2020
|
|
7EMD
 
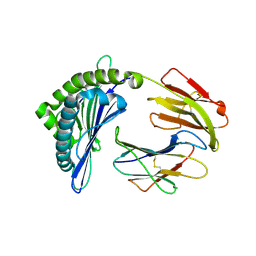 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, TYR-GLY-ASP-PHE-PHE-HIS-ASP-MET-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EM9
 
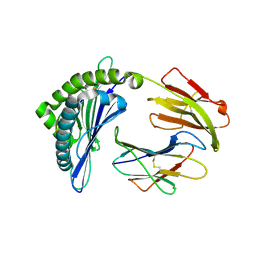 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, SER-LEU-ASP-GLU-TYR-SER-SER-ASP-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMA
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, TYR-SER-SER-ASP-VAL-THR-THR-LEU-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMB
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-ALA-ALA-ILE-GLU-GLU-GLU-ASP-ILE, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMC
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-THR-GLU-ILE-ARG-GLU-LEU-LEU-VAL, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
1X86
 
 | |
4MX5
 
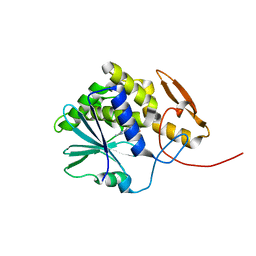 | | Structure of ricin A chain bound with benzyl-(2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)ethyl)carbamate | | Descriptor: | Ricin A chain, benzyl (2-{[(2-amino-4-oxo-3,4-dihydropteridin-7-yl)carbonyl]amino}ethyl)carbamate | | Authors: | Jasheway, K.R, Robertus, J.D, Wiget, P.A, Manzano, L.A, Pruet, J.M, Gao, G, Saito, R, Monzingo, A.F, Anslyn, E.V. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Sulfur incorporation generally improves Ricin inhibition in pterin-appended glycine-phenylalanine dipeptide mimics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MX1
 
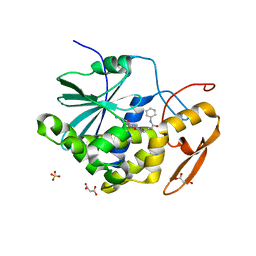 | | Structure of ricin A chain bound with 2-amino-4-oxo-N-(2-(3-phenylureido)ethyl)-3,4-dihydropteridine-7-carboxamide | | Descriptor: | 2-amino-4-oxo-N-{2-[(phenylcarbamoyl)amino]ethyl}-3,4-dihydropteridine-7-carboxamide, MALONIC ACID, Ricin A chain, ... | | Authors: | Robertus, J.D, Wiget, P.A, Manzano, L.A, Pruet, J.M, Gao, G, Saito, R, Jasheway, K.R, Monzingo, A.F, Anslyn, E.V. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sulfur incorporation generally improves Ricin inhibition in pterin-appended glycine-phenylalanine dipeptide mimics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6I2M
 
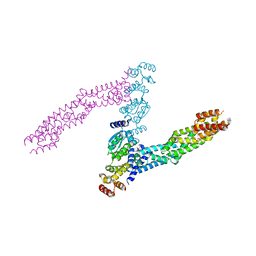 | |
4NFB
 
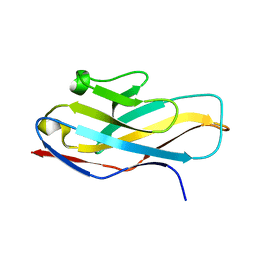 | | Structure of paired immunoglobulin-like type 2 receptor (PILR ) | | Descriptor: | Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7C01
 
 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | Authors: | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
6W3I
 
 | | Crystal structure of a FAM46C mutant in complex with Plk4 | | Descriptor: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | Authors: | Chen, H, Shang, G.J, Lu, D.F, Zhang, X.W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6W38
 
 | | Crystal structure of the FAM46C/Plk4 complex | | Descriptor: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | Authors: | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.48 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6W3J
 
 | | Crystal structure of the FAM46C/Plk4/Cep192 complex | | Descriptor: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | Authors: | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.385 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6W36
 
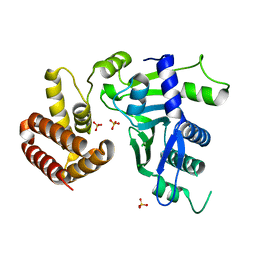 | | Crystal structure of FAM46C | | Descriptor: | SULFATE ION, Terminal nucleotidyltransferase 5C | | Authors: | Shang, G.J, Zhang, X.W, Chen, H, Lu, D.F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
