3BDK
 
 | | Crystal Structure of Streptococcus suis mannonate dehydratase complexed with substrate analogue | | Descriptor: | D-mannonate dehydratase, D-mannose, MANGANESE (II) ION | | Authors: | Gao, F, Zhang, Q.M, Peng, H, Liu, Y.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2007-11-15 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Streptococcus suis mannonate dehydratase complexed with substrate analogue
To be Published
|
|
8XAM
 
 | | Co-crystal structure of compound 7 in complex with MAT2A | | Descriptor: | 2-[3-[7-chloranyl-4-(dimethylamino)-2-oxidanylidene-quinazolin-1-yl]phenoxy]-~{N}-[3-[7-chloranyl-4-(dimethylamino)-2-oxidanylidene-quinazolin-1-yl]phenyl]ethanamide, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Gao, F, Ding, X. | | Deposit date: | 2023-12-04 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of novel MAT2A inhibitors by an allosteric site-compatible fragment growing approach.
Bioorg.Med.Chem., 100, 2024
|
|
8REB
 
 | |
8REA
 
 | |
8RE4
 
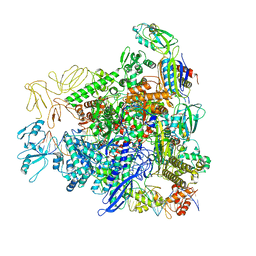 | |
8REC
 
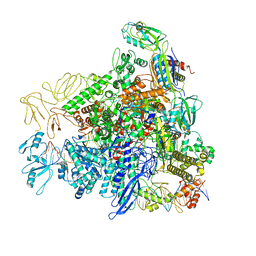 | |
8REE
 
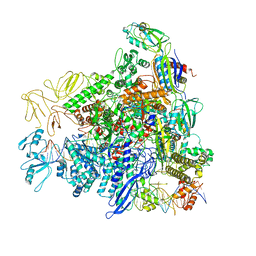 | |
8RED
 
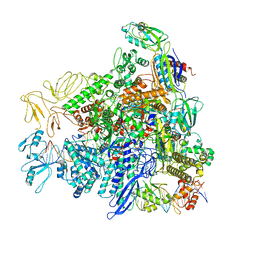 | |
7E78
 
 | | the structure of cytosolic TaPGI with substrate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Glucose-6-phosphate isomerase | | Authors: | Gao, F, Liu, C.M. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Engineering of the cytosolic form of phosphoglucose isomerase into chloroplasts improves plant photosynthesis and biomass.
New Phytol., 231, 2021
|
|
7E76
 
 | | The structure of chloroplastic TaPGI | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Gao, F, Liu, C.M. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineering of the cytosolic form of phosphoglucose isomerase into chloroplasts improves plant photosynthesis and biomass.
New Phytol., 231, 2021
|
|
7E77
 
 | | The structure of cytosolic TaPGI | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Gao, F, Liu, C.M. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Engineering of the cytosolic form of phosphoglucose isomerase into chloroplasts improves plant photosynthesis and biomass.
New Phytol., 231, 2021
|
|
2P5P
 
 | |
2OTP
 
 | | Crystal Structure of Immunoglobulin-Like Transcript 1 (ILT1/LIR7/LILRA2) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily A member 2 | | Authors: | Gao, F, Peng, H, Chen, Y, Liu, Y, Gao, G.F. | | Deposit date: | 2007-02-08 | | Release date: | 2008-02-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of 3D Domain Swapped Dimer of Immunoglobulin-Like Transcript 1 (ILT1/LIR7/LILRA2), Molecular Insight into Group 1 Activating Receptor Forming Unique MW Interaction Pattern
To be Published
|
|
3FAN
 
 | | Crystal structure of chymotrypsin-like protease/proteinase (3CLSP/Nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV) | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
3FAO
 
 | | Crystal structure of S118A mutant 3CLSP of PRRSV | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
7MXD
 
 | |
7N28
 
 | |
3BAN
 
 | | The crystal structure of mannonate dehydratase from Streptococcus suis serotype2 | | Descriptor: | D-mannonate dehydratase | | Authors: | Peng, H, Zhang, Q.M, Gao, F, Liu, Y.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2007-11-08 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of mannonate dehydratase from Streptococcus suis serotype2
To be Published
|
|
3CXR
 
 | | Crystal structure of gluconate 5-dehydrogase from streptococcus suis type 2 | | Descriptor: | Dehydrogenase with different specificities | | Authors: | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | Deposit date: | 2008-04-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the catalytic mechanism of gluconate 5-dehydrogenase from Streptococcus suis: Crystal structures of the substrate-free and quaternary complex enzymes.
Protein Sci., 18, 2009
|
|
3DBN
 
 | | Crystal structure of the Streptoccocus suis serotype2 D-mannonate dehydratase in complex with its substrate | | Descriptor: | D-MANNONIC ACID, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q, Gao, F, Gao, G.F. | | Deposit date: | 2008-06-02 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
4DFI
 
 | | Crystal structure of cell adhesion molecule nectin-2/CD112 mutant FAMP | | Descriptor: | Poliovirus receptor-related protein 2 | | Authors: | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2012-01-23 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
3I6K
 
 | | Newly identified epitope from SARS-CoV membrane protein complexed with HLA-A*0201 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Liu, J, Sun, Y, Qi, J, Chu, F, Wu, H, Gao, F, Li, T, Yan, J, Gao, G.F. | | Deposit date: | 2009-07-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes
J Infect Dis, 202, 2010
|
|
3FN3
 
 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
3MGT
 
 | | Crystal structure of a H5-specific CTL epitope variant derived from H5N1 influenza virus in complex with HLA-A*0201 | | Descriptor: | 10-meric peptide from Hemagglutinin, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Sun, Y, Liu, J, Yang, M, Gao, F, Zhou, J, Kitamura, Y. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses
J.Gen.Virol., 91, 2010
|
|
