3MNL
 
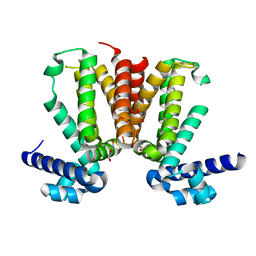 | | The crystal structure of KstR (Rv3574) from Mycobacterium tuberculosis H37Rv | | Descriptor: | TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY TETR-FAMILY), TRIETHYLENE GLYCOL | | Authors: | Gao, C, Bunker, R.D, ten Bokum, A, Kendall, S.L, Stoker, N.G, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-21 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J. Biol. Chem., 291, 2016
|
|
7DNR
 
 | |
7E88
 
 | |
7E86
 
 | |
7PHY
 
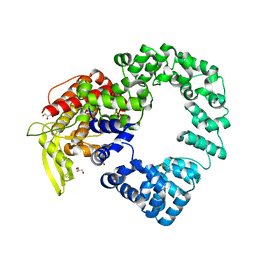 | | Vaccinia virus E2 | | Descriptor: | GLYCEROL, Protein E2 | | Authors: | Gao, W.N.D, Gao, C, Graham, S.C. | | Deposit date: | 2021-08-19 | | Release date: | 2021-09-01 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of vaccinia virus protein E2 and perspectives on the prediction of novel viral protein folds.
J.Gen.Virol., 103, 2022
|
|
3P8H
 
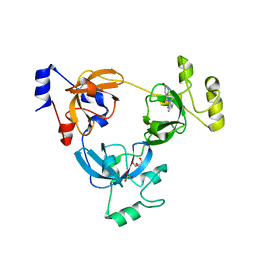 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | Descriptor: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | Authors: | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
3UWN
 
 | | The 3-MBT repeat domain of L3MBTL1 in complex with a methyl-lysine mimic | | Descriptor: | Lethal(3)malignant brain tumor-like protein 1, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | Authors: | Zhong, N, Tempel, W, Wernimont, A.K, Graslund, S, Ingerman, L.A, Korboukh, V, Kireev, D.B, Gao, C, Frye, S.V, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The 3-MBT repeat domain of L3MBTL1 in complex with a methyl-lysine mimic
To be Published
|
|
8IXI
 
 | |
6BL8
 
 | |
6K9L
 
 | | 4.27 Angstrom resolution cryo-EM structure of human dimeric ATM kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Chaban, Y, Gao, C, Tian, Y, Yu, Z, Li, J, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
6I2M
 
 | |
5BUO
 
 | | A receptor molecule | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETATE ION, Amyloid beta A4 protein, ... | | Authors: | Gao, C, Crespi, G.A.N, Gorman, M.A, Nero, T.L, Parker, M.W, Miles, L.A. | | Deposit date: | 2015-06-04 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | NULL
To Be Published
|
|
2QJ4
 
 | | A Mechanistic Basis for Converting a Receptor Tyrosine Kinase Agonist to an Antagonist | | Descriptor: | Hepatocyte growth factor, SULFATE ION | | Authors: | Tolbert, W.D, Daugherty, J, Gao, C.-F, Xe, Q, Miranti, C, Gherardi, E, Vande Woude, G, Xu, H.E. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QJ2
 
 | | A Mechanistic Basis for Converting a Receptor Tyrosine Kinase Agonist to an Antagonist | | Descriptor: | Hepatocyte growth factor, SULFATE ION | | Authors: | Tolbert, W.D, Daugherty, J, Gao, C.-F, Xe, Q, Miranti, C, Gherardi, E, Vande Woude, G, Xu, H.E. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7QDN
 
 | |
7QZU
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 47 | | Descriptor: | (2~{S})-2-[2-[4-[3,4-bis(oxidanyl)-9,10-bis(oxidanylidene)anthracen-2-yl]sulfonylpiperazin-1-yl]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2022-01-31 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
8HP3
 
 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
8HP0
 
 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, SULFATE ION | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
8HPE
 
 | | Crystal structure of Leucine dehydrogenase | | Descriptor: | GLYCEROL, Leucine dehydrogenase, SULFATE ION | | Authors: | Li, X, Song, W. | | Deposit date: | 2022-12-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Tri-Enzyme Cascade for Efficient Production of L-2-Aminobutyrate from L-Threonine.
Chembiochem, 24, 2023
|
|
8HR6
 
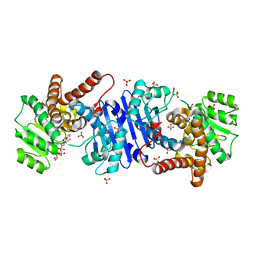 | |
7VVX
 
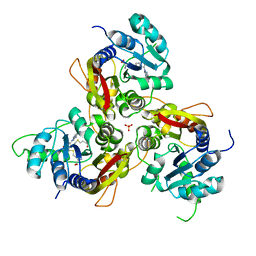 | | MmtN-SAH-Met complex | | Descriptor: | METHIONINE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVV
 
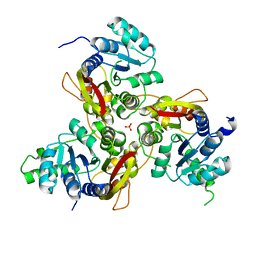 | | Crystal structure of MmtN | | Descriptor: | PHOSPHATE ION, SAM-dependent methyltransferase | | Authors: | Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVW
 
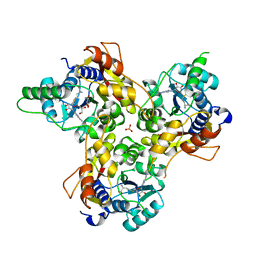 | | MmtN-SAM complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
3C2S
 
 | | Structural Characterization of a Human Fc Fragment Engineered for Lack of Effector Functions | | Descriptor: | IGHM protein, ZINC ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2008-01-25 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of a human Fc fragment engineered for lack of effector functions.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1Y5H
 
 | |
