2GJB
 
 | |
3T86
 
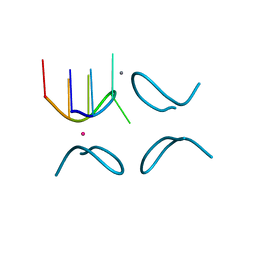 | | d(GCATGCT) + calcium | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), POTASSIUM ION | | Authors: | Cardin, C.J, Gan, Y. | | Deposit date: | 2011-08-01 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel structure for the d(GCATGCT) quadruplex in the presence of nickel and cobalt aqueous cations : comparison with the vanadium, barium and calcium-bound structural motif.
To be Published
|
|
1NQS
 
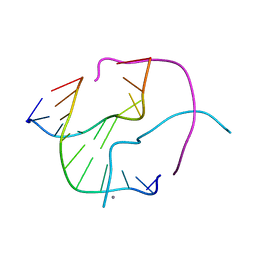 | | Structural Characterisation of the Holliday Junction formed by the sequence d(TCGGTACCGA) at 1.97 A | | Descriptor: | 5'-d(TpCpGpGpTpApCpCpGpA)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Texieira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-01-22 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Analysis of two Holliday junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
1NVN
 
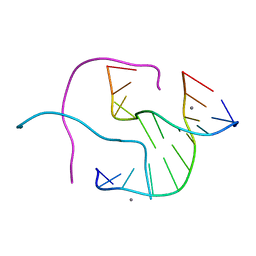 | | Structural Characterisation of the Holliday junction formed by the sequence CCGGTACCGG at 1.8 A | | Descriptor: | 5'-D(CpCpGpGpTpApCpCpGpG)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Teixeira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-02-04 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two Holliday junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
1NT8
 
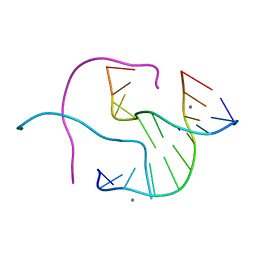 | | Structural Characterisation of the Holliday junction formed by the sequence CCGGTACCGG at 2.00 A | | Descriptor: | 5'-d(CpCpGpGpTpApCpCpGpG)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Texieira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-01-29 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of two Holliday Junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
1QYK
 
 | | GCATGCT + Barium | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*CP*T)-3', BARIUM ION | | Authors: | Cardin, C.J, Gan, Y, Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Moraes, M.I.A. | | Deposit date: | 2003-09-11 | | Release date: | 2003-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Metal Ion Distribution and Stabilization of the DNA Quadruplex Structure Formed by d(GCATGCT)
To be published
|
|
1QZL
 
 | | GCATGCT + Cobalt | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*CP*T)-3', COBALT (II) ION | | Authors: | Cardin, C.J, Gan, Y, Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Moraes, M.I.A. | | Deposit date: | 2003-09-17 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Metal Ion Distribution and Stabilisation of the DNA Quadruplex Structure Formed by d(GCATGCT)
To be published
|
|
1QYL
 
 | | GCATGCT + Vanadium | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*CP*T)-3', VANADIUM ION | | Authors: | Cardin, C.J, Gan, Y, Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Moraes, M.I.A. | | Deposit date: | 2003-09-11 | | Release date: | 2003-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Metal Ion Distribution and Stabilisation of the DNA Quadruplex Structure Formed by d(GCATGCT)
To be Published
|
|
1R2O
 
 | | d(GCATGCT) + Ni2+ | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*CP*T)-3', NICKEL (II) ION | | Authors: | Cardin, J.C, Gan, Y, Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Moraes, M.I.A. | | Deposit date: | 2003-09-29 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Metal Ion Distribution and Stabilization of the DNA Quadruplex Structure Formed by d(GCATGCT)
To be published
|
|
6V1N
 
 | | CSP1-E1A-cyc(Dap6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-01-08 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OLD
 
 | | CSP1-cyc(Dap6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-04-16 | | Release date: | 2020-01-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OC4
 
 | | CSP1-cyc(Dab6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OBW
 
 | | CSP1-cyc(K6D10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OC2
 
 | | CSP1-cyc(Orn6D10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6COW
 
 | | CSP1 | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COS
 
 | | CSP1-f11 | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COT
 
 | | CSP2-d10 | | Descriptor: | Competence-stimulating peptide type 2 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COV
 
 | | CSP2-l14 | | Descriptor: | Competence-stimulating peptide type 2 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COO
 
 | | CSP1-E1A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6CJ8
 
 | | CSP1 | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COP
 
 | | CSP1-R3A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COU
 
 | | CSP2-E1Ad10 | | Descriptor: | Competence-stimulating peptide type 2 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COQ
 
 | | CSP1-K6A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COR
 
 | | CSP1-F11A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6DDR
 
 | | Crystal Structure Analysis of the Epitope of an Anti-MICA Antibody | | Descriptor: | Anti-MICA Fab fragment heavy chain clone 13A9, Anti-MICA Fab fragment light chain clone 13A9, GLYCEROL, ... | | Authors: | Matsumoto, M.L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution glycosylation site-engineering method identifies MICA epitope critical for shedding inhibition activity of anti-MICA antibodies.
MAbs, 11, 2019
|
|
