4G34
 
 | | Crystal Structure of GSK6924 Bound to PERK (R587-R1092, delete A660-T867) at 2.70 A Resolution | | Descriptor: | 1-[5-(4-aminothieno[3,2-c]pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]-2-phenylethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
4G31
 
 | | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
4M7I
 
 | | Crystal Structure of GSK6157 Bound to PERK (R587-R1092, delete A660-T867) at 2.34A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-4-fluoro-1H-indol-1-yl]-2-(6-methylpyridin-2-yl)ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2013-08-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery of 5-{4-fluoro-1-[(6-methyl-2-pyridinyl)acetyl]-2,3-dihydro-1H-indol-5-yl}-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2656157), a Potent and Selective PERK Inhibitor Selected for Preclinical Development
To be Published
|
|
2Q7I
 
 | |
2Q7K
 
 | |
2Q7L
 
 | |
2Q7J
 
 | |
7JR6
 
 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1-(3-fluorophenyl)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7JR8
 
 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
2AUX
 
 | | Cathepsin K complexed with a semicarbazone inhibitor | | Descriptor: | (1R)-2-METHYL-1-(PHENYLMETHYL)PROPYL[(1S)-1-FORMYLPENTYL]CARBAMATE, Cathepsin K | | Authors: | Adkison, K.K, Barrett, D.G, Deaton, D.N, Gampe, R.T, Hassell, A.M, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Semicarbazone-based inhibitors of cathepsin K, are they prodrugs for aldehyde inhibitors?
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2AUZ
 
 | | Cathepsin K complexed with a semicarbazone inhibitor | | Descriptor: | 1-(PHENYLMETHYL)CYCLOPENTYL[(1S)-1-FORMYLPENTYL]CARBAMATE, Cathepsin K, SULFATE ION | | Authors: | Adkison, K.K, Barrett, D.G, Deaton, D.N, Gampe, R.T, Hassell, A.M, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Semicarbazone-based inhibitors of cathepsin K, are they prodrugs for aldehyde inhibitors?
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4Z2A
 
 | | Crystal structure of unglycosylated apo human furin @1.89A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Furin, ... | | Authors: | Gampe, R.T, Pearce, K, Reid, R. | | Deposit date: | 2015-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BacMam production and crystal structure of nonglycosylated apo human furin at 1.89A resolution
Acta Crystallogr.,Sect.F, 2019
|
|
3N00
 
 | |
6ZTC
 
 | | CRYSTAL STRUCTURE OF PROSTAGLANDIN D2 SYNTHASE IN COMPLEX WITH FRAGMENT 1A AT 1.84A RESOLUTION. | | Descriptor: | 1-[1-(3-fluorophenyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]propan-1-one, GLUTATHIONE, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-28 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
3L0E
 
 | | X-ray crystal structure of a Potent Liver X Receptor Modulator | | Descriptor: | N-(2-chloro-6-fluorobenzyl)-1-methyl-N-{[3'-(methylsulfonyl)biphenyl-4-yl]methyl}-1H-imidazole-4-sulfonamide, Nuclear receptor coactivator 2, Oxysterols receptor LXR-beta | | Authors: | Gampe Jr, R.T. | | Deposit date: | 2009-12-09 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of tertiary sulfonamides as potent liver X receptor antagonists.
J.Med.Chem., 53, 2010
|
|
3B3K
 
 | | Crystal structure of the complex between PPARgamma and the full agonist LT175 | | Descriptor: | (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Mazza, F, Loiodice, F, Fracchiolla, G, Crestani, M, Godio, C. | | Deposit date: | 2007-10-22 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Peroxisome Proliferator-Activated Receptor gamma (PPARgamma) Ligand Binding Domain Complexed with a Novel Partial Agonist: A New Region of the Hydrophobic Pocket Could Be Exploited for Drug Design
J.Med.Chem., 51, 2008
|
|
7LCU
 
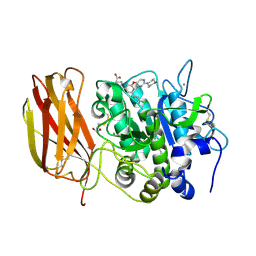 | | X-ray structure of Furin bound to BOS-318, a small molecule inhibitor | | Descriptor: | (1-{[2-(3,5-dichlorophenyl)-6-{[2-(4-methylpiperazin-1-yl)pyrimidin-5-yl]oxy}pyridin-4-yl]methyl}piperidin-4-yl)acetic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Campobasso, N, Reid, R. | | Deposit date: | 2021-01-11 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A highly selective, cell-permeable furin inhibitor BOS-318 rescues key features of cystic fibrosis airway disease.
Cell Chem Biol, 29, 2022
|
|
1KE8
 
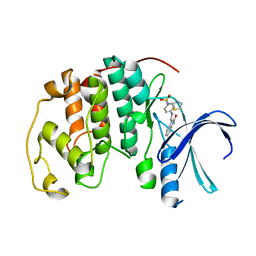 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE | | Descriptor: | 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE6
 
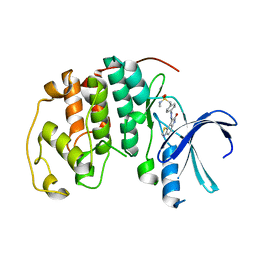 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Descriptor: | Cell division protein kinase 2, N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE9
 
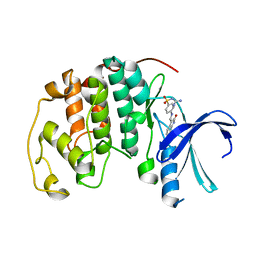 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[4-({[AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE | | Descriptor: | 3-{[4-([AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE7
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE | | Descriptor: | 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE5
 
 | | CDK2 complexed with N-methyl-4-{[(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]amino}benzenesulfonamide | | Descriptor: | Cell division protein kinase 2, N-METHYL-4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
3CDP
 
 | | Crystal structure of PPAR-gamma LBD complexed with a partial agonist, analogue of clofibric acid | | Descriptor: | (2S)-2-(4-chlorophenoxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-02-27 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, biological evaluation and molecular investigation of fluorinated peroxisome proliferator-activated receptors alpha/gamma dual agonists
Bioorg.Med.Chem., 20, 2012
|
|
3CDS
 
 | |
3D6D
 
 | | Crystal Structure of the complex between PPARgamma LBD and the LT175(R-enantiomer) | | Descriptor: | (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R. | | Deposit date: | 2008-05-19 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Peroxisome Proliferator-Activated Receptor gamma (PPARgamma) Ligand Binding Domain Complexed with a Novel Partial Agonist: A New Region of the Hydrophobic Pocket Could Be Exploited for Drug Design
J.Med.Chem., 51, 2008
|
|
