7Q0N
 
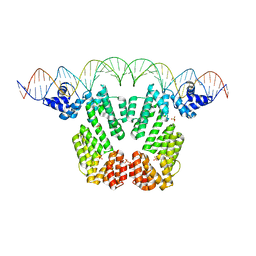 | | Arbitrium receptor from Katmira phage | | Descriptor: | Arbitrium receptor, DNA (45-MER), SULFATE ION | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the mechanism of action of the arbitrium communication system in SPbeta phages.
Nat Commun, 13, 2022
|
|
8BJ6
 
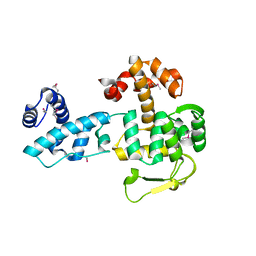 | | Crystal structure of YopR | | Descriptor: | SPbeta prophage-derived uncharacterized protein YopR | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of YopR from SPbeta phage
To Be Published
|
|
8BJV
 
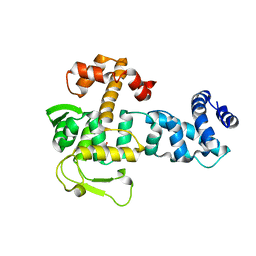 | | Crystal structure of YopR | | Descriptor: | GLYCEROL, SPbeta prophage-derived uncharacterized protein YopR | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YopR from SPbeta phage
To Be Published
|
|
8BPZ
 
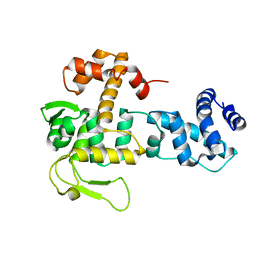 | | Crystal structure of YopR | | Descriptor: | SPbeta prophage-derived uncharacterized protein YopR | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YopR from SPbeta phage
To Be Published
|
|
1ZGR
 
 | |
1ZGS
 
 | |
2GDF
 
 | |
6S7I
 
 | |
6S7L
 
 | |
6HP5
 
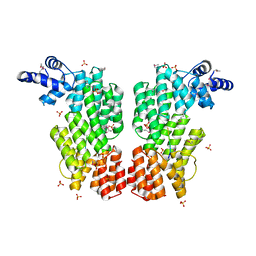 | | ARBITRIUM PEPTIDE RECEPTOR FROM SPBETA PHAGE | | Descriptor: | GLY-MET-PRO-ARG-GLY-ALA, SPBc2 prophage-derived uncharacterized protein YopK, SULFATE ION | | Authors: | Marina, A, Gallego del Sol, F. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems.
Mol.Cell, 74, 2019
|
|
6HP3
 
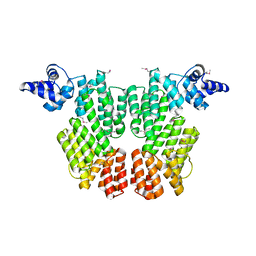 | |
6HP7
 
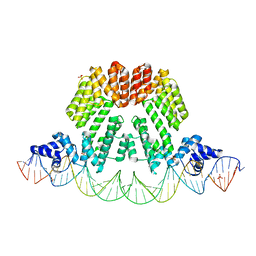 | |
4I9E
 
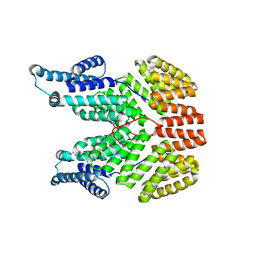 | |
4I9C
 
 | |
7ZTF
 
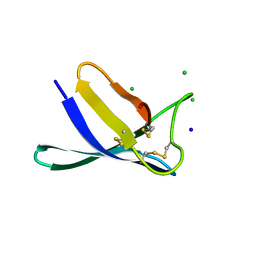 | | Penicillium expansum antifungal protein B | | Descriptor: | Antifungal protein, CHLORIDE ION, SODIUM ION | | Authors: | Gallego del Sol, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
7ZVH
 
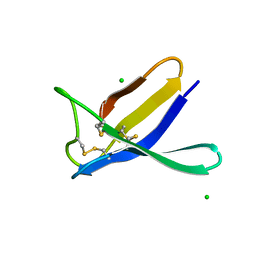 | | Penicillium expansum antifungal protein B | | Descriptor: | Antifungal protein, CHLORIDE ION | | Authors: | Gallego del Sol, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
7ZW2
 
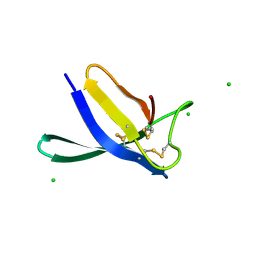 | | Penicillium expansum antifungal protein B | | Descriptor: | Antifungal protein, CHLORIDE ION | | Authors: | Gallego del Sol, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
1H9P
 
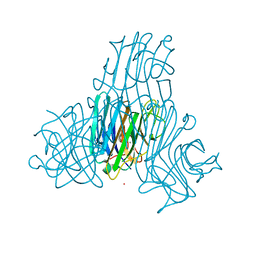 | | Crystal Structure of Dioclea guianensis Seed Lectin | | Descriptor: | CADMIUM ION, LECTIN ALPHA CHAIN, MANGANESE (II) ION | | Authors: | Romero, A, Wah, D.A, Gallego Del sol, F, Cavada, B.S, Ramos, M.V, Grangeiro, T.B, Sampaio, A.H, Calvete, J.J. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Native and Cd/Cd-Substituted Dioclea Guianensis Seed Lectin. A Novel Manganese-Binding Site and Structural Basis of Dimer-Tetramer Association
J.Mol.Biol., 310, 2001
|
|
