3IO5
 
 | |
4ZWQ
 
 | |
4ZWR
 
 | |
4ZWS
 
 | |
4ZWT
 
 | |
3SQ5
 
 | |
3SQ8
 
 | |
3SQ7
 
 | |
3SQ3
 
 | |
8GC7
 
 | |
8GC8
 
 | |
8EJB
 
 | |
8U2D
 
 | | Bruton's tyrosine kinase in complex with N-[(2R)-1-[(3R)-3-(methylcarbamoyl)-1H,2H,3H,4H,9H-pyrido[3,4-b]indol-2-yl]-3-(3-methylphenyl)-1-oxopropan-2-yl]-1H-indazole-5-carboxamide | | Descriptor: | (3R)-2-[N-(1H-indazole-5-carbonyl)-3-methyl-D-phenylalanyl]-N-methyl-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Gajewski, S, Clifton, M.C. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Preclinical Pharmacology of NX-2127, an Orally Bioavailable Degrader of Bruton's Tyrosine Kinase with Immunomodulatory Activity for the Treatment of Patients with B Cell Malignancies.
J.Med.Chem., 67, 2024
|
|
8U2E
 
 | | Bruton's tyrosine kinase in complex with N-[(2R)-1-[(3R)-3-(methylcarbamoyl)-1H,2H,3H,4H,9H-pyrido[3,4-b]indol-2-yl]-3-(3-methylphenyl)-1-oxopropan-2-yl]-1H-indazole-5-carboxamide | | Descriptor: | (2S)-6-fluoro-5-[(3S)-3-(3-methyl-2-oxoimidazolidin-1-yl)piperidin-1-yl]-2-(4-methylpiperazine-1-carbonyl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Gajewski, S, Clifton, M.C. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Preclinical Pharmacology of NX-2127, an Orally Bioavailable Degrader of Bruton's Tyrosine Kinase with Immunomodulatory Activity for the Treatment of Patients with B Cell Malignancies.
J.Med.Chem., 67, 2024
|
|
6CLU
 
 | |
6CLV
 
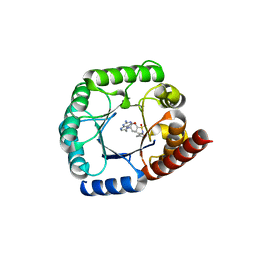 | | Staphylococcus aureus Dihydropteroate Synthase (saDHPS) F17L E208K double mutant structure | | Descriptor: | 4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-N-(3,4-dimethyl-1,2-oxazol-5-yl)benzene-1-sulfonamide, Dihydropteroate synthase | | Authors: | Gajewski, S, Griffith, E.C, Wu, Y, White, S.W. | | Deposit date: | 2018-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural and Functional Basis for Recurring Sulfa Drug Resistance Mutations inStaphylococcus aureusDihydropteroate Synthase.
Front Microbiol, 9, 2018
|
|
5KYM
 
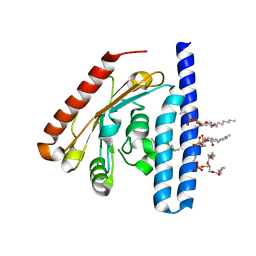 | | Crystal Structure of the 1-acyl-sn-glycerophosphate (LPA) acyltransferase, PlsC, from Thermotoga maritima | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, 1-acyl-sn-glycerol-3-phosphate acyltransferase, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Robertson, R.M, Yao, J, Gajewski, S, Kumar, G, Martin, E.W, Rock, C.O, White, S.W. | | Deposit date: | 2016-07-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A two-helix motif positions the lysophosphatidic acid acyltransferase active site for catalysis within the membrane bilayer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
