3UT2
 
 | | Crystal Structure of Fungal MagKatG2 | | Descriptor: | Catalase-peroxidase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zamocky, M, Garcia-Fernandez, M.Q, Gasselhuber, B, Jakopitsch, C, Furtmuller, P.G, Loewen, P.C, Fita, I, Obinger, C, Carpena, X. | | Deposit date: | 2011-11-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | High Conformational Stability of Secreted Eukaryotic Catalase-peroxidases: ANSWERS FROM FIRST CRYSTAL STRUCTURE AND UNFOLDING STUDIES.
J.Biol.Chem., 287, 2012
|
|
6XUB
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with monovinyl monopropionyl deuteroheme | | Descriptor: | Chlorite dismutase, harderoheme (III) | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
6XUC
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Chlorite dismutase | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8702 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
7Z53
 
 | |
7ATI
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 | | Descriptor: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
7ASB
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74E (CCld Q74E) from Cyanothece sp. PCC7425 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
4WWS
 
 | |
3QPI
 
 | | Crystal Structure of Dimeric Chlorite Dismutases from Nitrobacter winogradskyi | | Descriptor: | Chlorite Dismutase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mlynek, G, Sjoeblom, B, Kostan, J, Fuereder, S, Maixner, F, Furtmueller, P.G, Obinger, O, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2011-02-13 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradskyi.
J.Bacteriol., 193, 2011
|
|
6RWV
 
 | | Structure of apo-LmCpfC | | Descriptor: | Ferrochelatase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6386379 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
6SV3
 
 | | Structure of coproheme-LmCpfC | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Ferrochelatase, GLYCEROL | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-09-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64000869 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
5CJH
 
 | | Crystal Structure of Eukaryotic Oxoiron MagKatG2 at pH 8.5 | | Descriptor: | Catalase-peroxidase 2, HYDROXIDE ION, Peroxidized Heme | | Authors: | Gasselhuber, B, Obinger, C, Fita, I, Carpena, X. | | Deposit date: | 2015-07-14 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Eukaryotic Catalase-Peroxidase: The Role of the Trp-Tyr-Met Adduct in Protein Stability, Substrate Accessibility, and Catalysis of Hydrogen Peroxide Dismutation.
Biochemistry, 54, 2015
|
|
3F9P
 
 | | Crystal structure of myeloperoxidase from human leukocytes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Carpena, X, Fita, I, Obinger, C. | | Deposit date: | 2008-11-14 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Essential role of proximal histidine-asparagine interaction in Mammalian peroxidases.
J.Biol.Chem., 284, 2009
|
|
5MFA
 
 | | Crystal structure of human promyeloperoxidase (proMPO) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grishkovskaya, I, Furtmueller, P.G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human promyeloperoxidase (proMPO) and the role of the propeptide in processing and maturation.
J. Biol. Chem., 292, 2017
|
|
8OFL
 
 | | Coproporphyrin III - LmCpfC complex soaked 4min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Coproporphyrin III ferrochelatase, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
8OMM
 
 | | Coproporphyrin III - LmCpfC complex soaked 3min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, Coproporphyrin III ferrochelatase, FE (II) ION, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
8OGI
 
 | | Structure of native human eosinophil peroxidase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pfanzagl, V, Obinger, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Posttranslational modification and heme cavity architecture of human eosinophil peroxidase-insights from first crystal structure and biochemical characterization.
J.Biol.Chem., 299, 2023
|
|
6ERC
 
 | | Peroxidase A from Dictyostelium discoideum (DdPoxA) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nicolussi, A, Mlynek, G, Furtmueller, P.G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2017-10-17 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.50001836 Å) | | Cite: | Secreted heme peroxidase from Dictyostelium discoideum: Insights into catalysis, structure, and biological role.
J. Biol. Chem., 293, 2018
|
|
8BBV
 
 | | Coproporphyrin III - LmCpfC complex soaked 2min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Coproporphyrin III ferrochelatase, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
8AT8
 
 | | Structure of coproporphyrin III-LmCpfC | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Active site architecture of coproporphyrin ferrochelatase with its physiological substrate coproporphyrin III: Propionate interactions and porphyrin core deformation.
Protein Sci., 32, 2023
|
|
8AW7
 
 | | Structure of coproporphyrin III-LmCpfC R45L | | Descriptor: | Coproporphyrin III ferrochelatase, GLYCEROL, coproporphyrin III | | Authors: | Gabler, T, Hofbauer, S, Pfanzagl, V. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Active site architecture of coproporphyrin ferrochelatase with its physiological substrate coproporphyrin III: Propionate interactions and porphyrin core deformation.
Protein Sci., 32, 2023
|
|
5LOQ
 
 | | Structure of coproheme bound HemQ from Listeria monocytogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Puehringer, D, Mlynek, G, Hofbauer, S, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
FEBS J., 283, 2016
|
|
5MAU
 
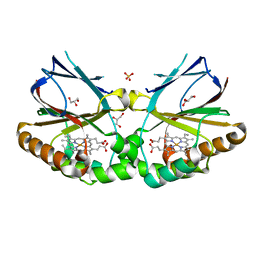 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 6.5) | | Descriptor: | Chlorite dismutase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5JHX
 
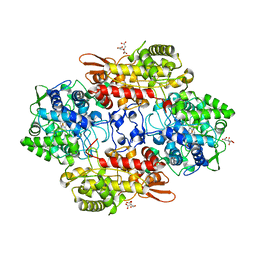 | | Crystal Structure of Fungal MagKatG2 at pH 3.0 | | Descriptor: | CITRATE ANION, Catalase-peroxidase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gasselhuber, B, Obinger, C, Carpena, X. | | Deposit date: | 2016-04-21 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interaction with the Redox Cofactor MYW and Functional Role of a Mobile Arginine in Eukaryotic Catalase-Peroxidase.
Biochemistry, 55, 2016
|
|
5JHY
 
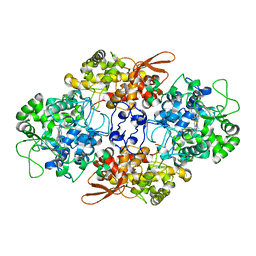 | |
5JHZ
 
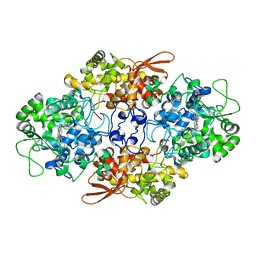 | |
