3LPB
 
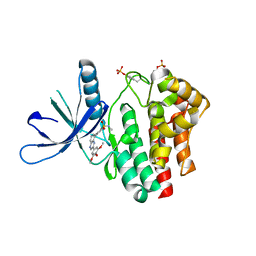 | | Crystal structure of Jak2 complexed with a potent 2,8-diaryl-quinoxaline inhibitor | | Descriptor: | N-methyl-4-[3-(3,4,5-trimethoxyphenyl)quinoxalin-5-yl]benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Tavares, G.A, Pissot-Soldermann, C, Gerspacher, M, Furet, P, Kroemer, M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and SAR of potent, orally available 2,8-diaryl-quinoxalines as a new class of JAK2 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4JPS
 
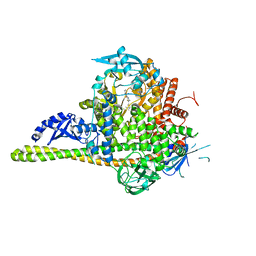 | | Co-crystal Structures of the Lipid Kinase PI3K alpha with Pan and Isoform Selective Inhibitors | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4DIJ
 
 | |
5LN2
 
 | |
8A0V
 
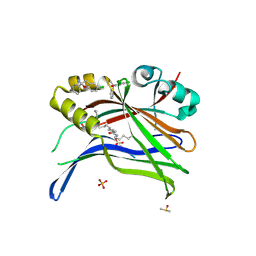 | | Crystal structure of TEAD3 in complex with CPD2 | | Descriptor: | DIMETHYL SULFOXIDE, MYRISTIC ACID, PHOSPHATE ION, ... | | Authors: | Scheufler, C, Kallen, J. | | Deposit date: | 2022-05-30 | | Release date: | 2022-08-17 | | Last modified: | 2022-10-19 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | The First Class of Small Molecules Potently Disrupting the YAP-TEAD Interaction by Direct Competition.
Chemmedchem, 17, 2022
|
|
8A0U
 
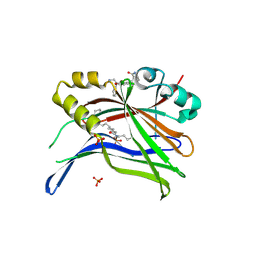 | | Crystal structure of TEAD3 in complex with CPD4 | | Descriptor: | 2-[(2~{S})-2-(aminomethyl)-5-chloranyl-2-phenyl-3~{H}-1-benzofuran-4-yl]benzamide, MYRISTIC ACID, PHOSPHATE ION, ... | | Authors: | Scheufler, C, Kallen, J. | | Deposit date: | 2022-05-30 | | Release date: | 2022-08-17 | | Last modified: | 2022-10-19 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | The First Class of Small Molecules Potently Disrupting the YAP-TEAD Interaction by Direct Competition.
Chemmedchem, 17, 2022
|
|
1CJ1
 
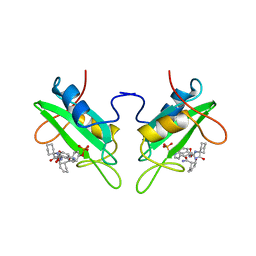 | | GROWTH FACTOR RECEPTOR BINDING PROTEIN SH2 DOMAIN (HUMAN) COMPLEXED WITH A PHOSPHOTYROSYL DERIVATIVE | | Descriptor: | PROTEIN (GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2), [1-[1-(6-CARBAMOYL-CYCLOHEX-2-ENYLCARBAMOYL)-CYCLOHEXYLCARBAMOYL]-2-(4-PHOSPHONOOXY-PHENYL)- ETHYL]-CARBAMIC ACID 3-AMINOBENZYLESTER | | Authors: | Rahuel, J. | | Deposit date: | 1999-04-21 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design, synthesis, and X-ray crystallography of a high-affinity antagonist of the Grb2-SH2 domain containing an asparagine mimetic.
J.Med.Chem., 42, 1999
|
|
1JVP
 
 | |
6Q36
 
 | |
6Q2X
 
 | |
5EW3
 
 | |
7TZ7
 
 | | PI3K alpha in complex with an inhibitor | | Descriptor: | (4S,5R)-3-[2'-amino-2-(morpholin-4-yl)-4'-(trifluoromethyl)[4,5'-bipyrimidin]-6-yl]-4-(hydroxymethyl)-5-methyl-1,3-oxazolidin-2-one, Isoform 3 of Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Tang, J. | | Deposit date: | 2022-02-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
8CAA
 
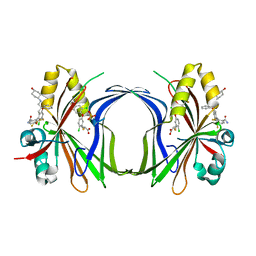 | | Crystal structure of TEAD4 in complex with YTP-13 | | Descriptor: | (2~{R})-2-[2-chloranyl-5-[2-chloranyl-4-(trifluoromethyl)phenoxy]phenyl]sulfanylpropanoic acid, 4-[bis(fluoranyl)methoxy]-2-[(2~{S})-5-chloranyl-6-fluoranyl-2-[[(4-oxidanylcyclohexyl)amino]methyl]-2-phenyl-3~{H}-1-benzofuran-4-yl]-3-fluoranyl-benzamide, PHOSPHATE ION, ... | | Authors: | Scheufler, C, Kallen, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-04-12 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Optimization of a Class of Dihydrobenzofurane Analogs toward Orally Efficacious YAP-TEAD Protein-Protein Interaction Inhibitors.
Chemmedchem, 18, 2023
|
|
3TT0
 
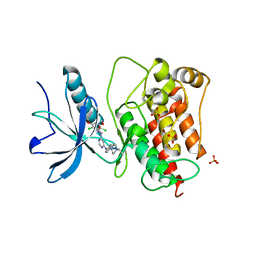 | | Co-structure of Fibroblast Growth Factor Receptor 1 kinase domain with 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (BGJ398) | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-(6-{[4-(4-ethylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1-methylurea, Basic fibroblast growth factor receptor 1, GLYCEROL, ... | | Authors: | Bussiere, D.E, Murray, J.M, Shu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase.
J.Med.Chem., 54, 2011
|
|
3KRR
 
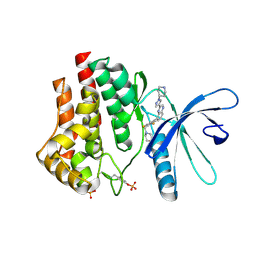 | | Crystal Structure of JAK2 complexed with a potent quinoxaline ATP site inhibitor | | Descriptor: | 8-[3,5-difluoro-4-(morpholin-4-ylmethyl)phenyl]-2-(1-piperidin-4-yl-1H-pyrazol-4-yl)quinoxaline, Tyrosine-protein kinase JAK2 | | Authors: | Tavares, G.A, Gerspacher, M, Kroemer, M, Scheufler, C. | | Deposit date: | 2009-11-19 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Inhibition of Polycythemia by the Quinoxaline JAK2 Inhibitor NVP-BSK805
Mol.Cancer Ther., 9, 2010
|
|
6Y20
 
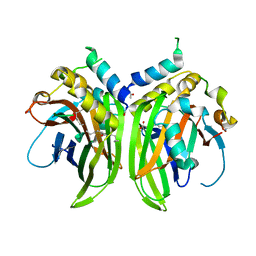 | | Crystal structure of Protein Scalloped (222-440) bound to Protein Vestigial (298-337) | | Descriptor: | AMINO GROUP, MYRISTIC ACID, Protein scalloped, ... | | Authors: | Scheufler, C, Villard, F, Bokhovchuk, F. | | Deposit date: | 2020-02-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | A new perspective on the interaction between the Vg/VGLL1-3 proteins and the TEAD transcription factors.
Sci Rep, 10, 2020
|
|
6YI8
 
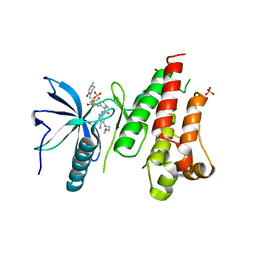 | | HUMAN FGFR4 KINASE DOMAIN (447-753) IN COMPLEX WITH ROBLITINIB | | Descriptor: | Fibroblast growth factor receptor 4, N-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-7-methanoyl-6-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3,4-dihydro-2H-1,8-naphthyridine-1-carboxamide, SULFATE ION | | Authors: | Ostermann, N. | | Deposit date: | 2020-04-01 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of Roblitinib (FGF401) as a Reversible-Covalent Inhibitor of the Kinase Activity of Fibroblast Growth Factor Receptor 4.
J.Med.Chem., 63, 2020
|
|
8P0M
 
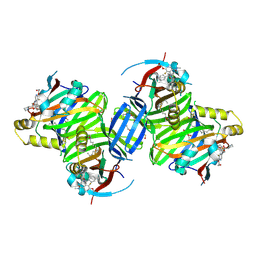 | | Crystal structure of TEAD3 in complex with IAG933 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-[(2~{S})-5-chloranyl-6-fluoranyl-2-phenyl-2-[(2~{S})-pyrrolidin-2-yl]-3~{H}-1-benzofuran-4-yl]-5-fluoranyl-6-(2-hydroxyethyloxy)-~{N}-methyl-pyridine-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Scheufler, C, Villard, F, Chau, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers.
Nat Cancer, 2024
|
|
4ZYC
 
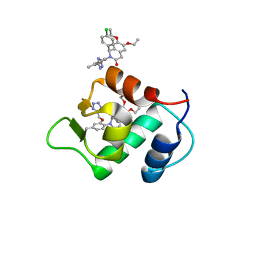 | |
4ZYF
 
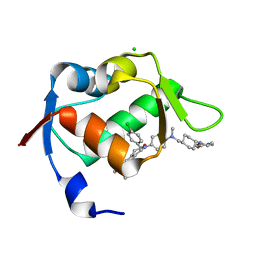 | |
4ZYI
 
 | |
6SEO
 
 | | TEAD4 bound to a FAM181B peptide | | Descriptor: | Protein FAM181B, Transcriptional enhancer factor TEF-3 | | Authors: | Scheufler, C, Villard, F. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of FAM181A and FAM181B as new interactors with the TEAD transcription factors.
Protein Sci., 29, 2020
|
|
6SEN
 
 | | TEAD4 bound to a FAM181A peptide | | Descriptor: | Protein FAM181A, SULFATE ION, Transcriptional enhancer factor TEF-3 | | Authors: | Scheufler, C, Villard, F. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of FAM181A and FAM181B as new interactors with the TEAD transcription factors.
Protein Sci., 29, 2020
|
|
3FEA
 
 | |
3FE7
 
 | |
