4PKF
 
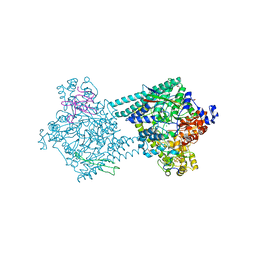 | | Benzylsuccinate synthase alpha-beta-gamma complex | | Descriptor: | CHLORIDE ION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structures of benzylsuccinate synthase elucidate roles of accessory subunits in glycyl radical enzyme activation and activity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PKC
 
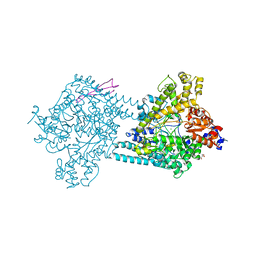 | | Benzylsuccinate alpha-gamma complex | | Descriptor: | CHLORIDE ION, GLYCEROL, TutD, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of benzylsuccinate synthase elucidate roles of accessory subunits in glycyl radical enzyme activation and activity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5BWE
 
 | |
5BWD
 
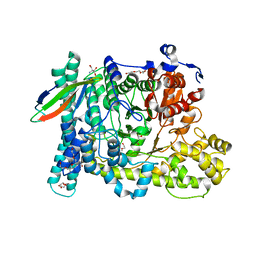 | | Benzylsuccinate alpha-gamma bound to fumarate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FUMARIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-06-07 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate-bound Structures of Benzylsuccinate Synthase Reveal How Toluene Is Activated in Anaerobic Hydrocarbon Degradation.
J.Biol.Chem., 290, 2015
|
|
4U3E
 
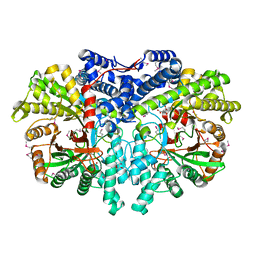 | | Anaerobic ribonucleotide reductase | | Descriptor: | ACETATE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-07-20 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The class III ribonucleotide reductase from Neisseria bacilliformis can utilize thioredoxin as a reductant.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5KDP
 
 | | E491A mutant of choline TMA-lyase | | Descriptor: | Choline trimethylamine-lyase, MALONATE ION, SODIUM ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
5FAV
 
 | | E491Q mutant of choline TMA-lyase | | Descriptor: | Choline trimethylamine-lyase, MALONATE ION, SODIUM ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-12-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
5FAY
 
 | | Y208F mutant of choline TMA-lyase | | Descriptor: | CHOLINE ION, Choline trimethylamine-lyase, MALONATE ION, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-12-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
5FAW
 
 | | T502A mutant of choline TMA-lyase | | Descriptor: | CHOLINE ION, Choline trimethylamine-lyase, MALONATE ION, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-12-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
5FAU
 
 | | wild-type choline TMA lyase in complex with choline | | Descriptor: | CHOLINE ION, Choline trimethylamine-lyase, GLYCEROL | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-12-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
6ND3
 
 | |
3D7M
 
 | | Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Funk, M.A, Preininger, A.M, Oldham, W.M, Meier, S.M, Hamm, H.E, Iverson, T.M. | | Deposit date: | 2008-05-21 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Helix dipole movement and conformational variability contribute to allosteric GDP release in Galphai subunits.
Biochemistry, 48, 2009
|
|
5CI4
 
 | | Ribonucleotide reductase beta subunit | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, beta subunit, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CI3
 
 | | Ribonucleotide reductase Y122 2,3,5-F3Y variant | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 subunit beta, SULFATE ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CI2
 
 | | Ribonucleotide reductase Y122 2,3,6-F3Y variant | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 subunit beta, SULFATE ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CI1
 
 | | Ribonucleotide reductase Y122 2,3-F2Y variant | | Descriptor: | CHLORIDE ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
5CI0
 
 | | Ribonucleotide reductase Y122 3,5-F2Y variant | | Descriptor: | MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1, beta subunit, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example.
J.Am.Chem.Soc., 138, 2016
|
|
6PO6
 
 | | MicroED Structure of a Natural Product VFAThiaGlu | | Descriptor: | YFAThiaGlu | | Authors: | Halaby, S, Gonen, T, Ting, C.P, Funk, M.A, van der Donk, W.A. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Use of a scaffold peptide in the biosynthesis of amino acid-derived natural products.
Science, 365, 2019
|
|
5CNV
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to GDP and TTP at 3.20 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
5CNS
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to CDP and dATP at 2.97 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.975 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
5CNT
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to UDP and dATP at 3.25 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, MU-OXO-DIIRON, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
5CNU
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to ADP and dGTP at 3.40 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
3UUS
 
 | |
3MXL
 
 | |
