2WZT
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
2WZM
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
3OAZ
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | CHLORIDE ION, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OAY
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OB0
 
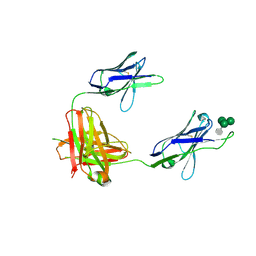 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | 7-deoxy-L-glycero-alpha-D-manno-heptopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OAU
 
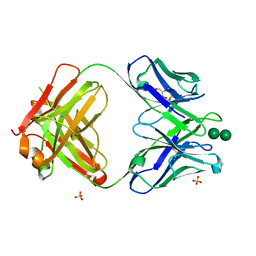 | | Antibody 2G12 Recognizes Di-Mannose Equivalently in Domain- and Non-Domain-Exchanged Forms, but only binds the HIV-1 Glycan Shield if Domain-Exchanged | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Huber, M, Wilson, I.A, Burton, D.R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged.
J.Virol., 84, 2010
|
|
3CKV
 
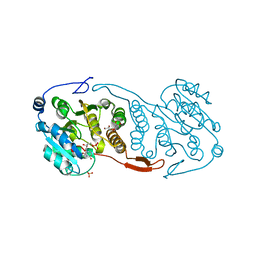 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKN
 
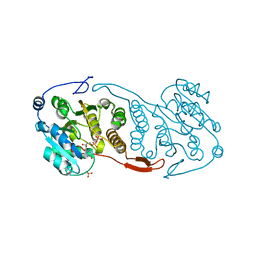 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | MANGANESE (II) ION, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKQ
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | MANGANESE (II) ION, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKJ
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CITRIC ACID, PHOSPHATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-15 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKO
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3BW9
 
 | | Crystal Structure of HLA B*3508 in complex with a HCMV 12-mer peptide from the pp65 protein | | Descriptor: | Beta-2-microglobulin, CPS peptide from 65 kDa lower matrix phosphoprotein, HLA class I histocompatibility antigen, ... | | Authors: | Wynn, K.K, Marland, Z, Cooper, L, Silins, S.L, Gras, S, Archbold, J.K, Tynan, F.E, Miles, J.J, McCluskey, J, Burrows, S.R, Rossjohn, J, Khanna, R. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Impact of clonal competition for peptide-MHC complexes on the CD8+ T-cell repertoire selection in a persistent viral infection
Blood, 111, 2008
|
|
3BWA
 
 | | Crystal Structure of HLA B*3508 in complex with a HCMV 8-mer peptide from the pp65 protein | | Descriptor: | Beta-2-microglobulin, FPT peptide from 65 kDa lower matrix phosphoprotein, HLA class I histocompatibility antigen, ... | | Authors: | Wynn, K.K, Marland, Z, Cooper, L, Silins, S.L, Gras, S, Archbold, J.K, Tynan, F.E, Miles, J.J, McCluskey, J, Burrows, S.R, Rossjohn, J, Khanna, R. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Impact of clonal competition for peptide-MHC complexes on the CD8+ T-cell repertoire selection in a persistent viral infection
Blood, 111, 2008
|
|
5V7U
 
 | |
5V7R
 
 | |
5N29
 
 | |
