4U4V
 
 | | Structure of a nitrate/nitrite antiporter NarK in apo inward-open state | | Descriptor: | NICKEL (II) ION, Nitrate/nitrite transporter NarK, OLEIC ACID | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4U4W
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4U4T
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound inward-open state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins
Cell(Cambridge,Mass.), 186, 2023
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins
Cell(Cambridge,Mass.), 186, 2023
|
|
8IU0
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins
Cell(Cambridge,Mass.), 186, 2023
|
|
1ULI
 
 | | Biphenyl dioxygenase (BphA1A2) derived from Rhodococcus sp. strain RHA1 | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, biphenyl dioxygenase large subunit, ... | | Authors: | Furusawa, Y, Nagarajan, V, Masai, E, Tanokura, M, Fukuda, M, Senda, T. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Biphenyl Dioxygenase Derived from Rhodococcus sp. Strain RHA1
J.Mol.Biol., 342, 2004
|
|
1ULJ
 
 | | Biphenyl dioxygenase (BphA1A2) in complex with the substrate | | Descriptor: | BIPHENYL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Furusawa, Y, Nagarajan, V, Masai, E, Tanokura, M, Fukuda, M, Senda, T. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Biphenyl Dioxygenase Derived from Rhodococcus sp. Strain RHA1
J.Mol.Biol., 342, 2004
|
|
1F3P
 
 | | FERREDOXIN REDUCTASE (BPHA4)-NADH COMPLEX | | Descriptor: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsuidagger, Y. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
2ZET
 
 | | Crystal structure of the small GTPase Rab27B complexed with the Slp homology domain of Slac2-a/melanophilin | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Melanophilin, ... | | Authors: | Kukimoto-Niino, M, Sakamoto, A, Kanno, E, Hanawa-Suetsugu, K, Terada, T, Shirouzu, M, Fukuda, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases.
Structure, 16, 2008
|
|
3A76
 
 | | The crystal structure of LinA | | Descriptor: | GLYCEROL, Gamma-hexachlorocyclohexane dehydrochlorinase, SPERMIDINE | | Authors: | Okai, M, Kubota, K, Fukuda, M, Nagata, Y, Nagata, K, Tanokura, M. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of g-hexachlorocyclohexane dehydrochlorinase LinA from Sphingobium japonicum UT26
J.Mol.Biol., 2010
|
|
7V73
 
 | | Thermostabilized human prestin in complex with chloride | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V75
 
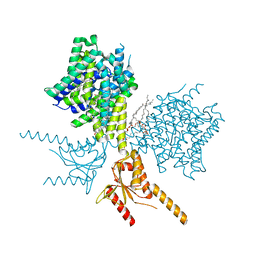 | | Thermostabilized human prestin in complex with salicylate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-HYDROXYBENZOIC ACID, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V74
 
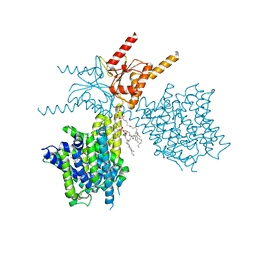 | | Thermostabilized human prestin in complex with sulfate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, SULFATE ION, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7C86
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: Dark state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RETINAL, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6Z
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 50 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6X
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 4 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E71
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6Y
 
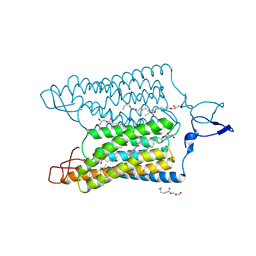 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E70
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 250 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7W9W
 
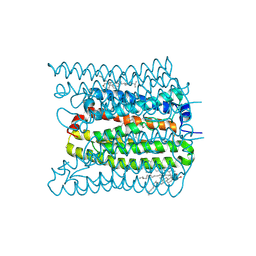 | | 2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine | | Descriptor: | CHOLESTEROL, ChRmine, PALMITIC ACID, ... | | Authors: | Kishi, K.E, Kim, Y, Fukuda, M, Yamashita, K, Deisseroth, K, Kato, H.E. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine.
Cell, 185, 2022
|
|
1D7Y
 
 | | CRYSTAL STRUCTURE OF NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4 | | Descriptor: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsui, Y. | | Deposit date: | 1999-10-21 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
2E4P
 
 | | Crystal structure of BphA3 (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER, SULFATE ION, ... | | Authors: | Senda, M, Kishigami, S, Kimura, S, Ishida, T, Fukuda, M, Senda, T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2E4Q
 
 | | Crystal structure of BphA3 (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Senda, M, Kishigami, S, Kimura, S, Ishida, T, Fukuda, M, Senda, T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
1BOU
 
 | | THREE-DIMENSIONAL STRUCTURE OF LIGAB | | Descriptor: | 4,5-DIOXYGENASE ALPHA CHAIN, 4,5-DIOXYGENASE BETA CHAIN, FE (III) ION | | Authors: | Sugimoto, K, Senda, T, Fukuda, M, Mitsui, Y. | | Deposit date: | 1998-08-06 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions.
Structure, 7, 1999
|
|
