1D9N
 
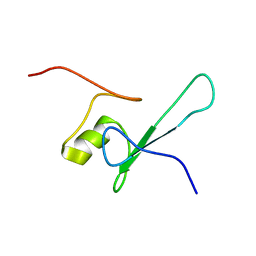 | | SOLUTION STRUCTURE OF THE METHYL-CPG-BINDING DOMAIN OF THE METHYLATION-DEPENDENT TRANSCRIPTIONAL REPRESSOR MBD1/PCM1 | | Descriptor: | METHYL-CPG-BINDING PROTEIN MBD1 | | Authors: | Ohki, I, Shimotake, N, Fujita, N, Nakao, M, Shirakawa, M. | | Deposit date: | 1999-10-28 | | Release date: | 2000-10-28 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1.
EMBO J., 18, 1999
|
|
1COO
 
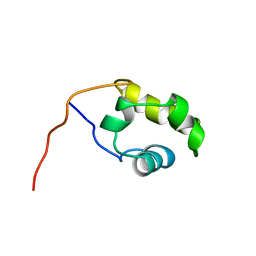 | | THE COOH-TERMINAL DOMAIN OF RNA POLYMERASE ALPHA SUBUNIT | | Descriptor: | RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Jeon, Y.H, Negishi, T, Shirakawa, M, Yamazaki, T, Fujita, N, Ishihama, A, Kyogoku, Y. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Science, 270, 1995
|
|
1IG4
 
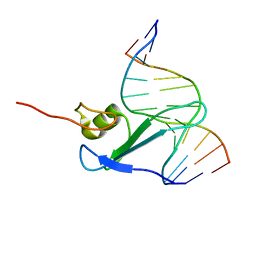 | | Solution Structure of the Methyl-CpG-Binding Domain of Human MBD1 in Complex with Methylated DNA | | Descriptor: | 5'-D(*GP*TP*AP*TP*CP*(5CM)P*GP*GP*AP*TP*AP*C)-3', Methyl-CpG Binding Protein | | Authors: | Ohki, I, Shimotake, N, Fujita, N, Jee, J.-G, Ikegami, T, Nakao, M, Shirakawa, M. | | Deposit date: | 2001-04-17 | | Release date: | 2001-05-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA.
Cell(Cambridge,Mass.), 105, 2001
|
|
3A99
 
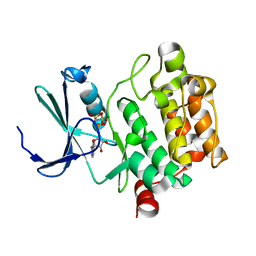 | | Structure of PIM-1 kinase crystallized in the presence of P27KIP1 Carboxy-terminal peptide | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Morishita, D, Takami, M, Yoshikawa, S, Katayama, R, Sato, S, Kukimoto-Niino, M, Umehara, T, Shirouzu, M, Sekimizu, K, Yokoyama, S, Fujita, N. | | Deposit date: | 2009-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cell-permeable carboxyl-terminal p27(Kip1) peptide exhibits anti-tumor activity by inhibiting Pim-1 kinase
J.Biol.Chem., 286, 2011
|
|
4MKC
 
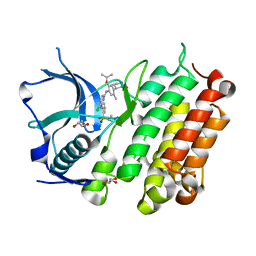 | | Crystal Structure of Anaplastic Lymphoma Kinase Complexed with LDK378 | | Descriptor: | 5-chloro-N~2~-[5-methyl-4-(piperidin-4-yl)-2-(propan-2-yloxy)phenyl]-N~4~-[2-(propan-2-ylsulfonyl)phenyl]pyrimidine-2,4-diamine, ALK tyrosine kinase receptor, GLYCEROL | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2013-09-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The ALK Inhibitor Ceritinib Overcomes Crizotinib Resistance in Non-Small Cell Lung Cancer.
Cancer Discov, 4, 2014
|
|
5GR0
 
 | |
5GQU
 
 | |
5GR6
 
 | |
5GR3
 
 | |
5GQV
 
 | |
5GR4
 
 | |
5GR1
 
 | |
5GQY
 
 | |
5GR2
 
 | |
5GR5
 
 | |
5GQX
 
 | |
5GQW
 
 | |
5GQZ
 
 | |
7A6C
 
 | |
7A65
 
 | |
7A6E
 
 | |
7A69
 
 | |
7A6F
 
 | |
8H7X
 
 | | Crystal structure of EGFR T790M/C797S mutant in complex with brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | A macrocyclic kinase inhibitor overcomes triple resistant mutations in EGFR-positive lung cancer.
NPJ Precis Oncol, 8, 2024
|
|
6KLF
 
 | |
