7ASS
 
 | |
6Q5F
 
 | | OXA-48_P68A-CAZ. Evolutionary trade-offs of OXA-48 mediated ceftazidime-avibactam resistance | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Frohlich, C, Sorum, V, Thomassen, A.M, Johnsen, P.J, Leiros, H.-K.S, Samuelsen, O. | | Deposit date: | 2018-12-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | OXA-48-Mediated Ceftazidime-Avibactam Resistance Is Associated with Evolutionary Trade-Offs.
Msphere, 4, 2019
|
|
6Q5B
 
 | | OXA-48_P68A-AVI. Evolutionary trade-offs of OXA-48 mediated ceftazidime-avibactam resistance | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, CARBON DIOXIDE, ... | | Authors: | Frohlich, C, Sorum, V, Thomassen, A.M, Johnsen, P.J, Leiros, H.K.S, Samuelsen, O. | | Deposit date: | 2018-12-07 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | OXA-48-Mediated Ceftazidime-Avibactam Resistance Is Associated with Evolutionary Trade-Offs.
Msphere, 4, 2019
|
|
6T5K
 
 | |
6T5L
 
 | |
8PEA
 
 | |
8PEC
 
 | |
8PEB
 
 | |
4BEJ
 
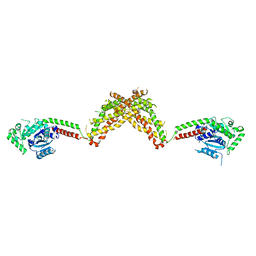 | | Nucleotide-free Dynamin 1-like protein (DNM1L, DRP1, DLP1) | | Descriptor: | DYNAMIN 1-LIKE PROTEIN | | Authors: | Froehlich, C, Schwefel, D, Faelber, K, Daumke, O. | | Deposit date: | 2013-03-11 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.483 Å) | | Cite: | Structural Insights Into Oligomerization and Mitochondrial Remodelling of Dynamin 1-Like Protein.
Embo J., 32, 2013
|
|
4H1V
 
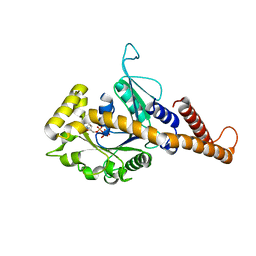 | | GMP-PNP bound dynamin-1-like protein GTPase-GED fusion | | Descriptor: | Dynamin-1-like protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Wenger, J, Klinglmayr, E, Eibl, C, Hessenberger, M, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
4H1U
 
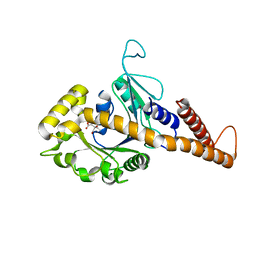 | | Nucleotide-free human dynamin-1-like protein GTPase-GED fusion | | Descriptor: | CITRATE ANION, Dynamin-1-like protein | | Authors: | Wenger, J, Klinglmayr, E, Puehringer, S, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
2XTN
 
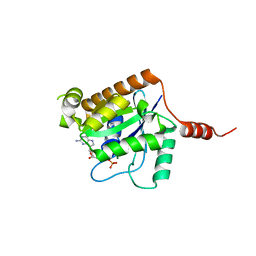 | | Crystal structure of GTP-bound human GIMAP2, amino acid residues 1- 234 | | Descriptor: | GTPASE IMAP FAMILY MEMBER 2, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Schwefel, D, Froehlich, C, Daumke, O. | | Deposit date: | 2010-10-11 | | Release date: | 2010-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Oligomerization in Septin-Like Gtpase of Immunity-Associated Protein 2 (Gimap2)
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XTM
 
 | | Crystal structure of GDP-bound human GIMAP2, amino acid residues 1- 234 | | Descriptor: | GLYCEROL, GTPASE IMAP FAMILY MEMBER 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schwefel, D, Froehlich, C, Daumke, O. | | Deposit date: | 2010-10-11 | | Release date: | 2010-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Oligomerization in Septin-Like Gtpase of Immunity-Associated Protein 2 (Gimap2)
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XTP
 
 | |
6TMB
 
 | |
6TMA
 
 | |
6TM9
 
 | |
6TMC
 
 | | VIM-2_1dh-Triazole inhibitors with promising inhibitor effects against antibiotic resistance metallo-beta-lactamases | | Descriptor: | 4-[2-(phenylsulfonyl)ethyl]-5-(propan-2-yloxymethyl)-1~{H}-1,2,3-triazole, Beta-lactamase class B VIM-2, HYDROXIDE ION, ... | | Authors: | Leiros, H.-K.S, Christopeit, T. | | Deposit date: | 2019-12-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies of triazole inhibitors with promising inhibitor effects against antibiotic resistance metallo-beta-lactamases.
Bioorg.Med.Chem., 28, 2020
|
|
