2WE0
 
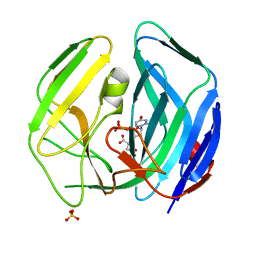 | | EBV dUTPase mutant Cys4Ser | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, MALATE LIKE INTERMEDIATE, ... | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
2WE1
 
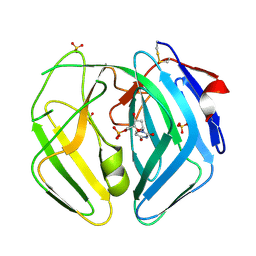 | | EBV dUTPase mutant Asp131Asn with bound dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, SULFATE ION | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
2WE2
 
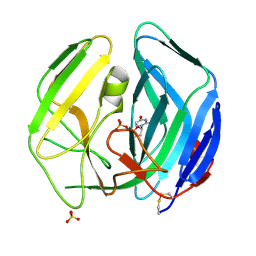 | | EBV dUTPase double mutant Gly78Asp-Asp131Ser with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, SULFATE ION | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
2WE3
 
 | | EBV dUTPase inactive mutant deleted of motif V | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, DEOXYURIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
2QZG
 
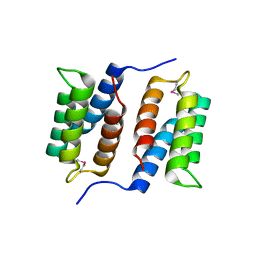 | | Crystal structure of unknown function protein MMP1188 | | Descriptor: | Conserved uncharacterized archaeal protein | | Authors: | Chang, C, Perez, V, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-16 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of MMP1188, unknown function protein.
To be Published
|
|
2R5R
 
 | | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718 | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, UPF0343 protein NE1163 | | Authors: | Tan, K, Wu, R, Nocek, B, Bigelow, L, Patterson, S, Freeman, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718.
To be Published
|
|
2R5S
 
 | | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Wu, R, Abdullah, J, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
3BOQ
 
 | |
3CJN
 
 | | Crystal structure of transcriptional regulator, MarR family, from Silicibacter pomeroyi | | Descriptor: | PHOSPHATE ION, Transcriptional regulator, MarR family | | Authors: | Chang, C, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MarR family transcriptional regulator from Silicibacter pomeroyi.
To be Published
|
|
3COL
 
 | |
3E6M
 
 | |
3N70
 
 | | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A | | Descriptor: | SULFATE ION, Transport activator | | Authors: | Stein, A.J, Mulligan, R, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A
To be Published
|
|
2QRR
 
 | | Crystal structure of the soluble domain of the ABC transporter, ATP-binding protein from Vibrio parahaemolyticus | | Descriptor: | CHLORIDE ION, Methionine import ATP-binding protein metN | | Authors: | Kim, Y, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-28 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Soluble Domain of the ABC Transporter, ATP-binding Protein from Vibrio parahaemolyticus.
To be Published
|
|
2QZI
 
 | | The crystal structure of a conserved protein of unknown function from Streptococcus thermophilus LMG 18311. | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | Authors: | Tan, K, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a conserved protein of unknown function from Streptococcus thermophilus LMG 18311.
To be Published
|
|
2QWV
 
 | | Crystal structure of unknown function protein VCA1059 | | Descriptor: | ACETIC ACID, UPF0217 protein VC_A1059 | | Authors: | Chang, C, Sather, A, Moy, S, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of unknown function protein VCA1059.
To be Published
|
|
3EEH
 
 | |
3LKL
 
 | | Crystal structure of the C-terminal domain of Anti-Sigma factor antagonist STAS from Rhodobacter sphaeroides | | Descriptor: | Antisigma-factor antagonist STAS | | Authors: | Nocek, B, Marshall, N, Davidoff, J, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the C-terminal domain of Anti-Sigma factor antagonist STAS from Rhodobacter sphaeroides
To be Published
|
|
3B49
 
 | |
3BHG
 
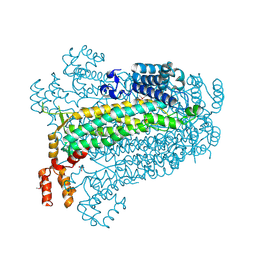 | | Crystal structure of adenylosuccinate lyase from Legionella pneumophila | | Descriptor: | Adenylosuccinate lyase, GLYCEROL, SULFATE ION | | Authors: | Chang, C, Li, H, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of adenylosuccinate lyase from Legionella pneumophila.
To be Published
|
|
3MXQ
 
 | |
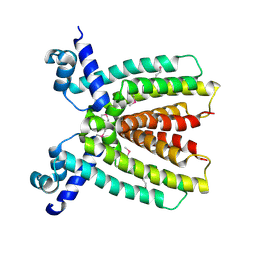
3BRU
 
 | |
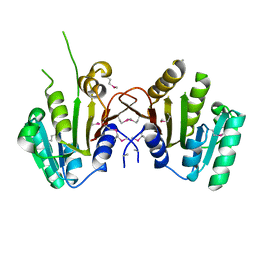
3MTI
 
 | |
3BWL
 
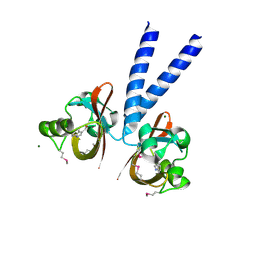 | | Crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, MAGNESIUM ION, Sensor protein | | Authors: | Osipiuk, J, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui.
To be Published
|
|
3NZN
 
 | | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1 | | Descriptor: | GLYCEROL, Glutaredoxin, SULFATE ION | | Authors: | Zhang, R, Wu, R, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1
To be Published
|
|
3OIZ
 
 | | Crystal structure of antisigma-factor antagonist, STAS domain from Rhodobacter sphaeroides | | Descriptor: | Antisigma-factor antagonist, STAS | | Authors: | Chang, C, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of antisigma-factor antagonist, STAS domain from Rhodobacter sphaeroides
To be Published
|
|
