4DNB
 
 | | METHYLATION OF THE ECORI RECOGNITION SITE DOES NOT ALTER DNA CONFORMATION. THE CRYSTAL STRUCTURE OF D(CGCGAM6ATTCGCG) AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*(6MA)P*TP*TP*CP*GP*CP*G)-3') | | Authors: | Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Methylation of the EcoRI recognition site does not alter DNA conformation: the crystal structure of d(CGCGAm6ATTCGCG) at 2.0-A resolution.
J.Biol.Chem., 263, 1988
|
|
1D13
 
 | | MOLECULAR STRUCTURE OF AN A-DNA DECAMER D(ACCGGCCGGT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Frederick, C.A, Quigley, G.J, Teng, M.-K, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of an A-DNA decamer d(ACCGGCCGGT).
Eur.J.Biochem., 181, 1989
|
|
1D10
 
 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
1D12
 
 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), DOXORUBICIN, SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
1AIO
 
 | | CRYSTAL STRUCTURE OF A DOUBLE-STRANDED DNA CONTAINING THE MAJOR ADDUCT OF THE ANTICANCER DRUG CISPLATIN | | Descriptor: | Cisplatin, DNA (5'-D(*CP*CP*(BRU)P*CP*TP*[PT(NH3)2(GP*GP)]*TP*CP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3') | | Authors: | Takahara, P.M, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 1997-04-23 | | Release date: | 1997-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin.
Nature, 377, 1995
|
|
1D17
 
 | | DNA-NOGALAMYCIN INTERACTIONS | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*AP*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Egli, M, Williams, L.D, Frederick, C.A, Rich, A. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-nogalamycin interactions.
Biochemistry, 30, 1991
|
|
1MTY
 
 | | METHANE MONOOXYGENASE HYDROXYLASE FROM METHYLOCOCCUS CAPSULATUS (BATH) | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Rosenzweig, A.C, Nordlund, P, Lippard, S.J, Frederick, C.A. | | Deposit date: | 1996-07-10 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions.
Proteins, 29, 1997
|
|
1CQK
 
 | | CRYSTAL STRUCTURE OF THE CH3 DOMAIN FROM THE MAK33 ANTIBODY | | Descriptor: | CH3 DOMAIN OF MAK33 ANTIBODY | | Authors: | Thies, M.J, Mayer, J, Augustine, J.G, Frederick, C.A, Lilie, H, Buchner, J. | | Deposit date: | 1999-08-06 | | Release date: | 1999-09-11 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Folding and association of the antibody domain CH3: prolyl isomerization preceeds dimerization.
J.Mol.Biol., 293, 1999
|
|
1D15
 
 | | TERNARY INTERACTIONS OF SPERMINE WITH DNA: 4'-EPIADRIAMYCIN AND OTHER DNA: ANTHRACYCLINE COMPLEXES | | Descriptor: | 4'-EPIDOXORUBICIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), SPERMINE | | Authors: | Williams, L.D, Frederick, C.A, Ughetto, G, Rich, A. | | Deposit date: | 1990-07-03 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ternary interactions of spermine with DNA: 4'-epiadriamycin and other DNA: anthracycline complexes.
Nucleic Acids Res., 18, 1990
|
|
1D14
 
 | | STRUCTURE OF 11-DEOXYDAUNOMYCIN BOUND TO DNA CONTAINING A PHOSPHOROTHIOATE | | Descriptor: | 6-DEOXYDAUNOMYCIN, DNA (5'-D(*CP*GP*TP*(AS)P*CP*G)-3') | | Authors: | Williams, L.D, Egli, M, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Quigley, G.J, Wang, A.H.-J, Rich, A, Frederick, C.A. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 11-deoxydaunomycin bound to DNA containing a phosphorothioate.
J.Mol.Biol., 215, 1990
|
|
1DCG
 
 | | THE MOLECULAR STRUCTURE OF THE LEFT-HANDED Z-DNA DOUBLE HELIX AT 1.0 ANGSTROM ATOMIC RESOLUTION. GEOMETRY, CONFORMATION, AND IONIC INTERACTIONS OF D(CGCGCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gessner, R.V, Frederick, C.A, Quigley, G.J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The molecular structure of the left-handed Z-DNA double helix at 1.0-A atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG).
J.Biol.Chem., 264, 1989
|
|
2ABL
 
 | |
2DND
 
 | | A BIFURCATED HYDROGEN-BONDED CONFORMATION IN THE D(A.T) BASE PAIRS OF THE DNA DODECAMER D(CGCAAATTTGCG) AND ITS COMPLEX WITH DISTAMYCIN | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Coll, M, Frederick, C.A, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin.
Proc.Natl.Acad.Sci.USA, 84, 1987
|
|
1LAR
 
 | | CRYSTAL STRUCTURE OF THE TANDEM PHOSPHATASE DOMAINS OF RPTP LAR | | Descriptor: | PROTEIN (LAR) | | Authors: | Nam, H.-J, Poy, F, Krueger, N, Saito, H, Frederick, C.A. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the tandem phosphatase domains of RPTP LAR.
Cell(Cambridge,Mass.), 97, 1999
|
|
1QBP
 
 | |
1MMO
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL NON-HAEM IRON HYDROXYLASE THAT CATALYSES THE BIOLOGICAL OXIDATION OF METHANE | | Descriptor: | ACETIC ACID, FE (III) ION, METHANE MONOOXYGENASE HYDROLASE (ALPHA CHAIN), ... | | Authors: | Rosenzweig, A.C, Frederick, C.A, Lippard, S.J, Nordlund, P. | | Deposit date: | 1994-02-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane.
Nature, 366, 1993
|
|
1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
1DNH
 
 | | THE MOLECULAR STRUCTURE OF THE COMPLEX OF HOECHST 33258 AND THE DNA DODECAMER D(CGCGAATTCGCG) | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Teng, M.-K, Usman, N, Frederick, C.A, Wang, A.H.-J. | | Deposit date: | 1988-02-16 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG).
Nucleic Acids Res., 16, 1988
|
|
1FH5
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MONOCLONAL ANTIBODY MAK33 | | Descriptor: | MONOCLONAL ANTIBODY MAK33 | | Authors: | Augustine, J.G, de la Calle, A, Knarr, G, Buchner, J, Frederick, C.A. | | Deposit date: | 2000-07-31 | | Release date: | 2000-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the fab fragment of the monoclonal antibody MAK33. Implications for folding and interaction with the chaperone bip.
J.Biol.Chem., 276, 2001
|
|
1FZI
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM I PRESSURIZED WITH XENON GAS | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ALPHA CHAIN, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
1FZ9
 
 | | Methane monooxygenase hydroxylase, form II cocrystallized with iodoethane | | Descriptor: | CALCIUM ION, FE (III) ION, IODOETHANE, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
1FZH
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM II PRESSURIZED WITH XENON GAS | | Descriptor: | CALCIUM ION, FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
1FZ8
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM II COCRYSTALLIZED WITH DIBROMOMETHANE | | Descriptor: | CALCIUM ION, DIBROMOMETHANE, FE (III) ION, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
1VTT
 
 | | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*G)-3') | | Authors: | Ho, P.S, Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG)
Embo J., 4, 1985
|
|
1YGR
 
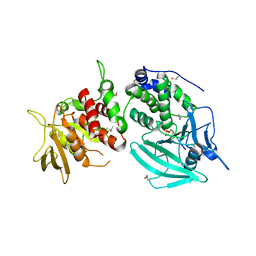 | | Crystal structure of the tandem phosphatase domain of RPTP CD45 | | Descriptor: | CD45 Protein Tyrosine Phosphatase, T-cell Receptor CD3 zeta ITAM-1 | | Authors: | Nam, H.J, Poy, F, Saito, H, Frederick, C.A. | | Deposit date: | 2005-01-05 | | Release date: | 2005-02-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the function and regulation of the receptor protein tyrosine phosphatase CD45.
J.Exp.Med., 201, 2005
|
|
