3TGP
 
 | | Room temperature H-ras | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Fraser, J.S, Alber, T. | | Deposit date: | 2011-08-17 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3075 Å) | | Cite: | Accessing protein conformational ensembles using room-temperature X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K0P
 
 | |
3K0M
 
 | | Cryogenic structure of CypA | | Descriptor: | Cyclophilin A | | Authors: | Fraser, J.S, Alber, T. | | Deposit date: | 2009-09-24 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hidden alternative structures of proline isomerase essential for catalysis.
Nature, 462, 2009
|
|
3K0R
 
 | |
3K0N
 
 | | Room temperature structure of CypA | | Descriptor: | Cyclophilin A | | Authors: | Fraser, J.S, Alber, T. | | Deposit date: | 2009-09-24 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Hidden alternative structures of proline isomerase essential for catalysis.
Nature, 462, 2009
|
|
3K0Q
 
 | |
3K0O
 
 | |
2NT3
 
 | | Receiver domain from Myxococcus xanthus social motility protein FrzS (Y102A Mutant) | | Descriptor: | Response regulator homolog | | Authors: | Fraser, J.S, Echols, N, Merlie, J.P, Zusman, D.R, Alber, T. | | Deposit date: | 2006-11-06 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
5F66
 
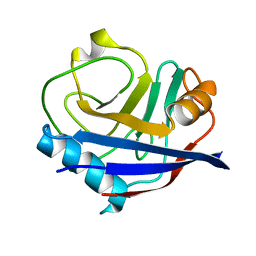 | |
7RGR
 
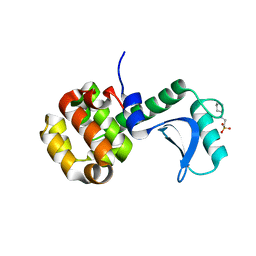 | | Lysozyme 056 from Deep neural language modeling | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Artificial protein L056, CHLORIDE ION | | Authors: | Fraser, J.S, Holton, J.M, Olmos Jr, J.L, Greene, E.R. | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Large language models generate functional protein sequences across diverse families.
Nat.Biotechnol., 2023
|
|
5WC7
 
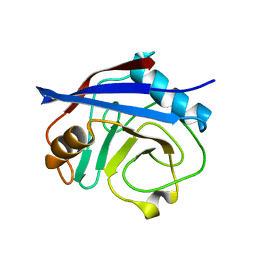 | | CypA Mutant - I97V S99T C115S | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Fraser, J.S. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Rescue of conformational dynamics in enzyme catalysis by directed evolution.
Nat Commun, 9, 2018
|
|
6BTA
 
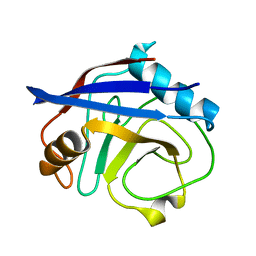 | | CypA Mutant - S99T C115S | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Fraser, J.S, Kenner, L.R, Liu, L. | | Deposit date: | 2017-12-06 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescue of conformational dynamics in enzyme catalysis by directed evolution.
Nat Commun, 9, 2018
|
|
4OBV
 
 | | Ruminococcus gnavus tryptophan decarboxylase RUMGNA_01526 (alpha-FMT) | | Descriptor: | Pyridoxal-dependent decarboxylase domain protein, alpha-(fluoromethyl)-D-tryptophan, {5-hydroxy-4-[(1E)-4-(1H-indol-3-yl)-3-oxobut-1-en-1-yl]-6-methylpyridin-3-yl}methyl dihydrogen phosphate | | Authors: | Fraser, J.S, Van Benschoten, A.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine.
Cell Host Microbe, 16, 2014
|
|
6UAD
 
 | | Ketosteroid isomerase (C. testosteroni) with truncated & designed loop for precise positioning of a catalytic E38 | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Ketosteroid isomerase with truncated and designed loop, PHOSPHATE ION | | Authors: | Kundert, K, Thompson, M.C, Liu, L, Fraser, J.S, Kortemme, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ketosteroid isomerase (C. testosteroni) with truncated & designed loop for precise positioning of a catalytic E38
To Be Published
|
|
6DMH
 
 | | A multiconformer ligand model of acylenzyme intermediate of meropenem bound to an SFC-1 E166A mutant | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6DMK
 
 | | A multiconformer ligand model of an isoxazolyl-benzimidazole ligand bound to the bromodomain of human CREBBP | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazole, CREB-binding protein, ... | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
5KUZ
 
 | | Human cyclophilin A at 278K, Data set 1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KW0
 
 | | T. danielli thaumatin at 100K, Data set 5 | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-15 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXX
 
 | | Hen Egg White Lysozyme at 278K, Data set 7 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KUR
 
 | | Human cyclophilin A at 100K, Data set 5 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KVX
 
 | | T. danielli thaumatin at 100K, Data set 2 | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-15 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXO
 
 | | Hen Egg White Lysozyme at 278K, Data set 1 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXW
 
 | | Hen Egg White Lysozyme at 278K, Data set 6 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KV6
 
 | | Human cyclophilin A at 278K, Data set 8 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXZ
 
 | | Hen Egg White Lysozyme at 278K, Data set 9 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
