2BEE
 
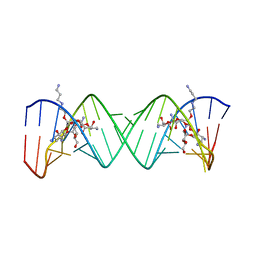 | | Complex Between Paromomycin derivative JS4 and the 16S-Rrna A Site | | Descriptor: | (2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3R,4R,5S)-4-(2-(3-AMINOPROPYLAMINO)ETHOXY)-5-((1R,2R,3S,5R,6S)-3,5-DIAM INO-2-((2S,3R,4R,5S,6R)-3-AMINO-4,5-DIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-2-( HYDROXYMETHYL)-TETRAHYDROFURAN-3-YLOXY)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Francois, B, Westhof, E. | | Deposit date: | 2005-10-24 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site
ANGEW.CHEM.INT.ED.ENGL., 43, 2004
|
|
2BE0
 
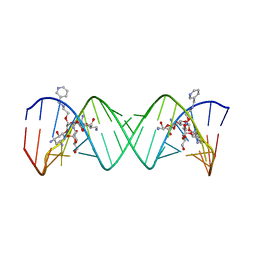 | | Complex Between Paromomycin Derivative JS5-39 and the 16S-Rrna A-Site. | | Descriptor: | (2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3R,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R,4R,5S,6R)-3-AMINO-4 ,5-DIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-2-(HYDROXYMETHYL)-4-(2-((R)-PIPERIDI N-3-YLMETHYLAMINO)ETHOXY)-TETRAHYDROFURAN-3-YLOXY)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Francois, B, Westhof, E. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site
ANGEW.CHEM.INT.ED.ENGL., 43, 2004
|
|
2PWT
 
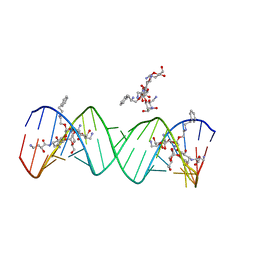 | | Crystal structure of the bacterial ribosomal decoding site complexed with aminoglycoside containing the L-HABA group | | Descriptor: | 22-mer of the ribosomal decoding site, DOUBLY FUNCTIONALIZED PAROMOMYCIN PM-II-162 | | Authors: | Kondo, J, Pachamuthu, K, Francois, B, Szychowski, J, Hanessian, S, Westhof, E. | | Deposit date: | 2007-05-13 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Bacterial Ribosomal Decoding Site Complexed with a Synthetic Doubly Functionalized Paromomycin Derivative: a New Specific Binding Mode to an A-Minor Motif Enhances in vitro Antibacterial Activity
Chemmedchem, 2, 2007
|
|
4P20
 
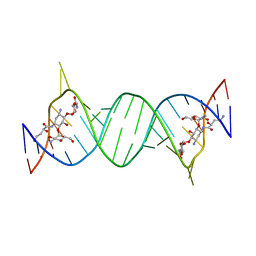 | | Crystal structures of the bacterial ribosomal decoding site complexed with amikacin | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Kondo, J, Francois, B, Russell, R.J.M, Murray, J.B, Westhof, E. | | Deposit date: | 2014-02-28 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the bacterial ribosomal decoding site complexed with amikacin containing the gamma-amino-alpha-hydroxybutyryl (haba) group.
Biochimie, 88, 2006
|
|
2G5K
 
 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with Apramycin | | Descriptor: | 5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3', APRAMYCIN, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Francois, B, Urzhumtsev, A, Westhof, E. | | Deposit date: | 2006-02-23 | | Release date: | 2006-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site Complexed with Apramycin
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2ET5
 
 | | Complex Between Ribostamycin and the 16S-RRNA A-Site | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3', RIBOSTAMYCIN, SULFATE ION | | Authors: | Westhof, E. | | Deposit date: | 2005-10-27 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding.
Nucleic Acids Res., 33, 2005
|
|
2ET8
 
 | | Complex Between Neamine and the 16S-RRNA A-Site | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Westhof, E. | | Deposit date: | 2005-10-27 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding.
Nucleic Acids Res., 33, 2005
|
|
2ESI
 
 | | Complex between Kanamycin A and the 16S-Rrna A Site. | | Descriptor: | 5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3', KANAMYCIN A | | Authors: | Westhof, E. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding
Nucleic Acids Res., 33, 2005
|
|
2ET3
 
 | | Complex Between Gentamicin C1A and the 16S-RRNA A-Site | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Westhof, E. | | Deposit date: | 2005-10-27 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding.
Nucleic Acids Res., 33, 2005
|
|
2ESJ
 
 | | Complex between Lividomycin A and the 16S-Rrna A Site | | Descriptor: | (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Westhof, E. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding
Nucleic Acids Res., 33, 2005
|
|
2ET4
 
 | | Complex Between Neomycin B and the 16S-RRNA A-Site | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3', NEOMYCIN | | Authors: | Westhof, E. | | Deposit date: | 2005-10-27 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding.
Nucleic Acids Res., 33, 2005
|
|
