1A3M
 
 | | PAROMOMYCIN BINDING INDUCES A LOCAL CONFORMATIONAL CHANGE IN THE A SITE OF 16S RRNA, NMR, 20 STRUCTURES | | Descriptor: | 16S RRNA (5'-R(*GP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*UP*UP*C)-3'), 16S RRNA (5'-R(*GP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*C)-3') | | Authors: | Fourmy, D, Yoshizawa, S, Puglisi, J.D. | | Deposit date: | 1998-01-22 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA.
J.Mol.Biol., 277, 1998
|
|
1MED
 
 | |
1MEA
 
 | |
1PBR
 
 | | STRUCTURE OF 16S RIBOSOMAL RNA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 16S RIBOSOMAL RNA, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXYSTREPTAMINE, ... | | Authors: | Fourmy, D, Recht, M.I, Blanchard, S, Puglisi, J.D. | | Deposit date: | 1996-09-12 | | Release date: | 1997-09-17 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic.
Science, 274, 1996
|
|
1MFK
 
 | | Structure of Prokaryotic SECIS mRNA Hairpin | | Descriptor: | 5'-R(P*GP*GP*CP*GP*GP*UP*UP*GP*CP*AP*GP*GP*UP*CP*UP*GP*CP*AP*CP*CP*GP*CP*C)-3' | | Authors: | Fourmy, D, Guittet, E, Yoshizawa, S. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of Prokaryotic SECIS mRNA Hairpin and its Interaction with Elongation Factor SELB
J.Mol.Biol., 324, 2002
|
|
1BYJ
 
 | | GENTAMICIN C1A A-SITE COMPLEX | | Descriptor: | 2,6-diamino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranose, 3,5-DIAMINO-CYCLOHEXANOL, 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranose, ... | | Authors: | Yoshizawa, S, Fourmy, D, Puglisi, J.D. | | Deposit date: | 1998-10-16 | | Release date: | 1999-10-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural origins of gentamicin antibiotic action.
EMBO J., 17, 1998
|
|
1WSU
 
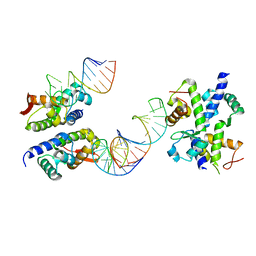 | | C-terminal domain of elongation factor selB complexed with SECIS RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*GP*CP*CP*GP*GP*UP*CP*U*GP*GP*CP*AP*AP*CP*GP*CP*C)-3', Selenocysteine-specific elongation factor | | Authors: | Yoshizawa, S, Rasubala, L, Ose, T, Kohda, D, Fourmy, D, Maenaka, K. | | Deposit date: | 2004-11-11 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for mRNA recognition by elongation factor SelB
Nat.Struct.Mol.Biol., 12, 2005
|
|
2PLY
 
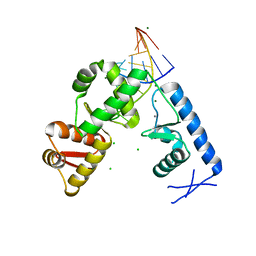 | | Structure of the mRNA binding fragment of elongation factor SelB in complex with SECIS RNA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Soler, N, Fourmy, D, Yoshizawa, S. | | Deposit date: | 2007-04-20 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB.
J.Mol.Biol., 370, 2007
|
|
2PJP
 
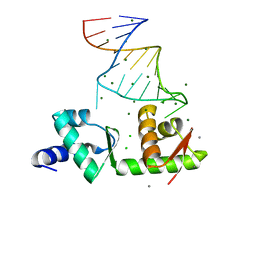 | | Structure of the mRNA-binding domain of elongation factor SelB from E.coli in complex with SECIS RNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Soler, N, Fourmy, D, Yoshizawa, S. | | Deposit date: | 2007-04-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB.
J.Mol.Biol., 370, 2007
|
|
2UWM
 
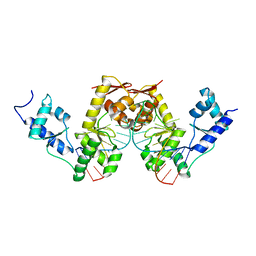 | | C-TERMINAL DOMAIN(WH2-WH4) OF ELONGATION FACTOR SELB IN COMPLEX WITH SECIS RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*GP*CP*CP*GP *GP*UP*CP*UP*GP*GP*CP*AP*AP*CP*GP*CP*C)-3', SELENOCYSTEINE-SPECIFIC ELONGATION FACTOR | | Authors: | Ose, T, Soler, N, Rasubala, L, Kuroki, K, Kohda, D, Fourmy, D, Yoshizawa, S, Maenaka, K. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Dynamic Interdomain Movement and RNA Recognition of the Selenocysteine-Specific Elongation Factor Selb.
Structure, 15, 2007
|
|
1ZC5
 
 | | Structure of the RNA signal essential for translational frameshifting in HIV-1 | | Descriptor: | HIV-1 frameshift RNA signal | | Authors: | Gaudin, C, Mazauric, M.H, Traikia, M, Guittet, E, Yoshizawa, S, Fourmy, D. | | Deposit date: | 2005-04-11 | | Release date: | 2005-06-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the RNA Signal Essential for Translational Frameshifting in HIV-1
J.Mol.Biol., 349, 2005
|
|
2GV3
 
 | | Translocation of a tRNA with an extended anticodon through the ribosome | | Descriptor: | 5'-R(*GP*GP*CP*CP*AP*GP*AP*CP*UP*CP*CP*CP*GP*AP*AP*UP*CP*UP*GP*GP*CP*C)-3' | | Authors: | Phelps, S.S, Gaudin, C, Yoshizawa, S, Benitez, C, Fourmy, D, Joseph, S. | | Deposit date: | 2006-05-02 | | Release date: | 2006-08-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Translocation of a tRNA with an Extended Anticodon Through the Ribosome.
J.Mol.Biol., 360, 2006
|
|
1K6H
 
 | | Solution Structure of Conserved AGNN Tetraloops: Insights into Rnt1p RNA processing | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*UP*GP*UP*UP*CP*AP*GP*AP*AP*GP*AP*AP*CP*GP*CP*GP*CP*C)-3') | | Authors: | Lebars, I, Lamontagne, B, Yoshizawa, S, Abou Elela, S, Fourmy, D. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing.
EMBO J., 20, 2001
|
|
1K6G
 
 | | Solution Structure of Conserved AGNN Tetraloops: Insights into Rnt1p RNA Processing | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*UP*CP*AP*UP*GP*AP*GP*UP*CP*CP*AP*UP*GP*GP*CP*GP*CP*C)-3') | | Authors: | Lebars, I, Lamontagne, B, Yoshizawa, S, Abou Elela, S, Fourmy, D. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing.
EMBO J., 20, 2001
|
|
1MT4
 
 | | Structure of 23S ribosomal RNA hairpin 35 | | Descriptor: | 23S ribosomal Hairpin 35 | | Authors: | Lebars, I, Yoshizawa, S, Stenholm, A.R, Guittet, E, Douthwaite, S, Fourmy, D. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of 23S rRNA hairpin 35 and its interaction with the tylosin-resistance methyltransferase RlmAII
Embo J., 22, 2003
|
|
2HNS
 
 | | Structure of the AAGU tetraloop | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*GP*AP*UP*CP*AP*AP*GP*UP*GP*AP*UP*CP*GP*CP*GP*CP*C)-3' | | Authors: | Gaudin, C, Fourmy, F, Yoshizawa, S. | | Deposit date: | 2006-07-13 | | Release date: | 2006-10-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of an AAGU Tetraloop and its Contribution to Substrate Selection by yeast RNase III.
J.Mol.Biol., 363, 2006
|
|
