7LQK
 
 | | Crystal structure of the R375A mutant of LeuT | | Descriptor: | ALANINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Font, J, Aguilar, J, Galli, A, Ryan, R. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Psychomotor impairments and therapeutic implications revealed by a mutation associated with infantile Parkinsonism-Dystonia.
Elife, 10, 2021
|
|
7LQJ
 
 | | Crystal structure of LeuT bound to L-Alanine | | Descriptor: | ALANINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Font, J, Aguilar, J, Galli, A, Ryan, R. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Psychomotor impairments and therapeutic implications revealed by a mutation associated with infantile Parkinsonism-Dystonia.
Elife, 10, 2021
|
|
7LQL
 
 | | Crystal structure of the R375D mutant of LeuT | | Descriptor: | ALANINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Font, J, Aguilar, J, Galli, A, Ryan, R. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Psychomotor impairments and therapeutic implications revealed by a mutation associated with infantile Parkinsonism-Dystonia.
Elife, 10, 2021
|
|
6WYJ
 
 | | Cryo-EM structure of the GltPh L152C-G321C mutant in the intermediate state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog | | Authors: | Font, J, Chen, I, Sobti, M, Stewart, A.G, Ryan, R.M. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
6WYL
 
 | | Cryo-EM structure of GltPh L152C-G351C mutant in the intermediate outward-facing state. | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog | | Authors: | Font, J, Chen, I, Sobti, M, Stewart, A.G, Ryan, R.M. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
6WYK
 
 | | Cryo-EM structure of the GltPh L152C-G321C mutant in the intermediate chloride conducting state. | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog | | Authors: | Font, J, Chen, I, Sobti, M, Stewart, A.G, Ryan, R.M. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
6BAU
 
 | | Crystal Structure of GltPh R397C in complex with L-Cysteine | | Descriptor: | CYSTEINE, Glutamate transporter homolog, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-10-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
6BAT
 
 | | Crystal Structure of Wild-Type GltPh in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-10-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
6BAV
 
 | | Crystal Structure of GltPh R397C in complex with S-Benzyl-L-Cysteine | | Descriptor: | BENZYLCYSTEINE, Glutamate transporter homolog, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
6BMI
 
 | | Crystal Structure of GltPh R397C in complex with L-Serine | | Descriptor: | Glutamate transporter homolog, SERINE, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
6X01
 
 | |
6WZB
 
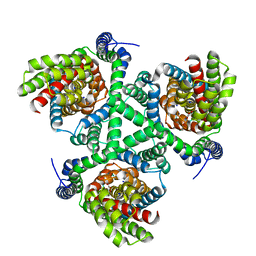 | | Crystal structure of the GltPh V216C-G388C mutant cross-linked with divalent mercury | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, MERCURY (II) ION, ... | | Authors: | Chen, I, Font, J, Ryan, R. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
6HYE
 
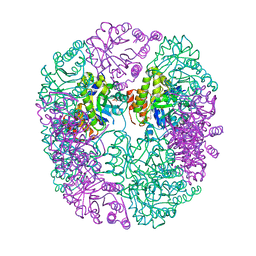 | | PDX1.2/PDX1.3 complex (PDX1.3:K97A) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6HXG
 
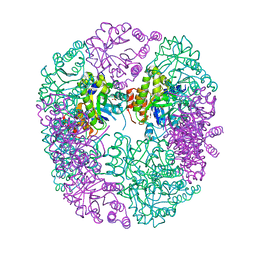 | | PDX1.2/PDX1.3 complex (intermediate) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6HX3
 
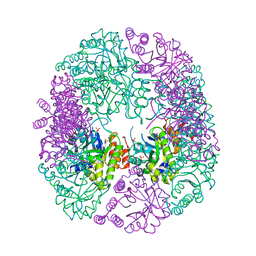 | | PDX1.2/PDX1.3 complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3DH6
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (V47A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3DI9
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (I81A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3DI8
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (V57A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3DH5
 
 | | Crystal structure of bovine pancreatic ribonuclease A (wild-type) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3DI7
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (V54A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3DIB
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (I106A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3DIC
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (V108A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
5AYN
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, Solute carrier family 39 (Iron-regulated transporter) | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
5AYO
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in inward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, Solute carrier family 39 (Iron-regulated transporter), ... | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
5AYM
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in outward-facing state with soaked iron | | Descriptor: | FE (II) ION, Solute carrier family 39 (Iron-regulated transporter) | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
