2VLD
 
 | | crystal structure of a repair endonuclease from Pyrococcus abyssi | | Descriptor: | Endonuclease NucS | | Authors: | Ren, B, Kuhn, J, Meslet-Cladiere, L, Briffotaux, J, Norais, C, Lavigne, R, Flament, D, Ladenstein, R, Myllykallio, H. | | Deposit date: | 2008-01-14 | | Release date: | 2009-05-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of a novel endonuclease acting on branched DNA substrates.
EMBO J., 28, 2009
|
|
4HR1
 
 | | Structure of PAV1-137, a protein from the virus PAV1 that infects Pyrococcus abyssi. | | Descriptor: | Putative uncharacterized protein | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, Geslin, C, Flament, D, Romancer, M.L, Tilbeurgh, H.V. | | Deposit date: | 2012-10-26 | | Release date: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of PAV1-137: a protein from the virus PAV1 that infects Pyrococcus abyssi
Archaea, 2013, 2013
|
|
1YRA
 
 | | PAB0955 crystal structure : a GTPase in GDP bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
2OXR
 
 | | PAB0955 crystal structure : a GTPase in GDP and Mg bound form from Pyrococcus abyssi (after GTP hydrolysis) | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
7ANU
 
 | | Crystal structure of Pyrococcus abyssi Pbp11 | | Descriptor: | GLYCEROL, IMIDAZOLE, NICKEL (II) ION, ... | | Authors: | Hogrel, G, Girard, E, Franzetti, B. | | Deposit date: | 2020-10-12 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization of a small tRNA-binding protein that interacts with the archaeal proteasome complex.
Mol.Microbiol., 118, 2022
|
|
5EQT
 
 | |
1YR6
 
 | |
1YR9
 
 | | PAB0955 crystal structure : a GTPase in GDP and PO4 bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1YR7
 
 | | PAB0955 crystal structure : a GTPase in GTP-gamma-S bound form from Pyrococcus abyssi | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ATP(GTP)binding protein | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1YR8
 
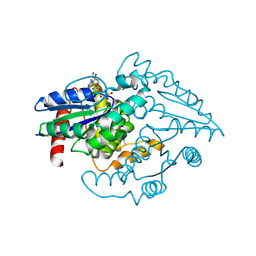 | | PAB0955 crystal structure : a GTPase in GTP bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1YRB
 
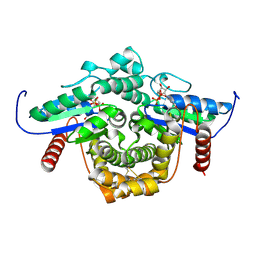 | | PAB0955 crystal structure : a GTPase in GDP and Mg bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
8OEJ
 
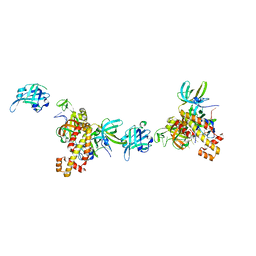 | | Extended RPA-DNA nucleoprotein filament | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Sauguet, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (7.96 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8OEL
 
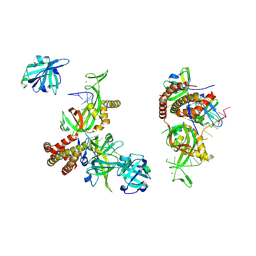 | | Condensed RPA-DNA nucleoprotein filament | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Sauguet, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (8.24 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8C5Y
 
 | | RPA tetrameric supercomplex from Pyrococcus abyssi | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Legrand, P, Sauguet, L. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8C5Z
 
 | | RPA tetrameric supercomplex with AROD-OB-1 | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Legrand, P, Sauguet, L. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8AAJ
 
 | | Crystal structure of the Pyrococcus abyssi RPA (apo form) | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Legrand, P, Madru, C, Sauguet, L. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8AAS
 
 | | Crystal structure of the Pyrococcus abyssi RPA trimerization core bound to poly-dT20 ssDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, ... | | Authors: | Madru, C, Legrand, P, Sauguet, L. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8AA9
 
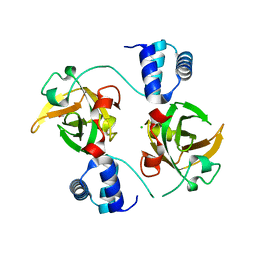 | |
7OUE
 
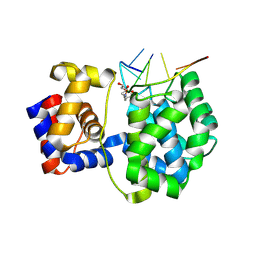 | | Crystal structure of a trapped Pab-AGOG/single-standed DNA covalent intermediate | | Descriptor: | DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), N-glycosylase/DNA lyase, PHOSPHATE ION, ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OY7
 
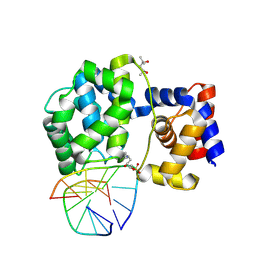 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing cytosine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
1DYP
 
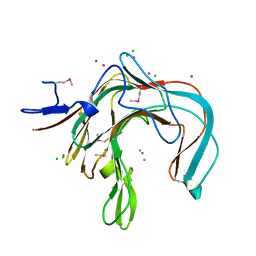 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE-3,6-ANHYDRO-D-GALACTOSE 4 GALACTOHYDROLASE | | Descriptor: | CADMIUM ION, CHLORIDE ION, KAPPA-CARRAGEENASE | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-02-04 | | Release date: | 2001-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Kappa-Carrageenase of P. Carrageenovora Features a Tunnel-Shaped Active Site: A Novel Insight in the Evolution of Clan-B Glycoside Hydrolases
Structure, 9, 2001
|
|
1H80
 
 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE- 3,6-ANHYDRO-D-GALACTOSE-2-SULFATE 4 GALACTOHYDROLASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2001-01-22 | | Release date: | 2001-11-27 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Iota-Carrageenase of Alteromonas Fortis. A Beta-Helix Fold-Containing Enzyme for the Degradation of a Highly Polyanionic Polysaccharide
J.Biol.Chem., 276, 2001
|
|
7OLI
 
 | | Crystal structure of Pab-AGOG in complex with 8-oxoguanosine | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-8-OXOGUANOSINE, N-glycosylase/DNA lyase | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P0W
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing thymine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OLB
 
 | | Crystal structure of Pab-AGOG, an 8-oxoguanine DNA glycosylase from Pyrococcus abyssi | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
