4CFI
 
 | | 3D structure of FliC from Burkholderia pseudomallei | | Descriptor: | FLAGELLIN | | Authors: | Lassaux, P, Peri, C, Ferrer-Navarro, M, Gourlay, L.J, Conchillo-Sole, O, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sequence- and Structure-Based Immunoreactive Epitope Discovery for Burkholderia Pseudomallei Flagellin.
Plos Negl Trop Dis, 9, 2015
|
|
4B5C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia pseudomallei | | Descriptor: | ACETATE ION, PUTATIVE OMPA FAMILY LIPOPROTEIN | | Authors: | Gourlay, L.J, Peri, C, Conchillo-Sole, O, Ferrer-Navarro, M, Gori, A, Longhi, R, Rinchai, D, Lertmemongkolchai, G, Lassaux, P, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Burkholderia Pseudomallei Acute Phase Antigen Bpsl2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology.
Chem.Biool., 20, 2013
|
|
4BWR
 
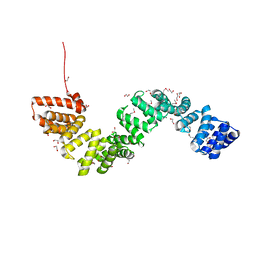 | | Crystal structure of c5321: a protective antigen present in uropathogenic Escherichia coli strains displaying an SLR fold | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Urosev, D, Ferrer-Navarro, M, Pastorello, I, Cartocci, E, Costenaro, L, Zhulenkovs, D, Marechal, J.-D, Leonchiks, A, Reverter, D, Serino, L, Soriani, M, Daura, X. | | Deposit date: | 2013-07-04 | | Release date: | 2013-07-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of C5321: A Protective Antigen Present in Uropathogenic Escherichia Coli Strains Displaying an Slr Fold.
Bmc Struct.Biol., 13, 2013
|
|
4UT1
 
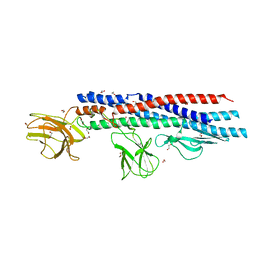 | |
3N26
 
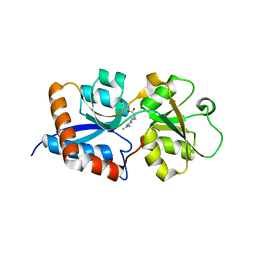 | | Cpn0482 : the arginine binding protein from the periplasm of chlamydia Pneumoniae | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Petit, P, Garcia, C, Vuillard, L, Soriani, M, Grandi, G, Marseilles Structural Genomics Program AFMB (MSGP), Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploiting antigenic diversity for vaccine design: the Chlamydia ArtJ paradigm.
J.Biol.Chem., 2010
|
|
3DEL
 
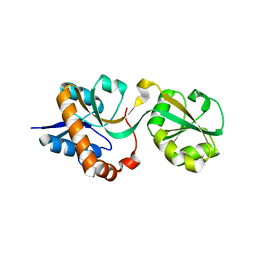 | |
6HSY
 
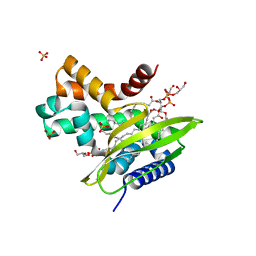 | | Two-phospholipid-bound crystal structure of the substrate-binding protein Ttg2D from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Phospholipid PG(16:0/cy17:0), SULFATE ION, ... | | Authors: | Costenaro, L, Conchillo-Sole, O, Daura, X. | | Deposit date: | 2018-10-02 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The Pseudomonas aeruginosa substrate-binding protein Ttg2D functions as a general glycerophospholipid transporter across the periplasm.
Commun Biol, 4, 2021
|
|
3ZS6
 
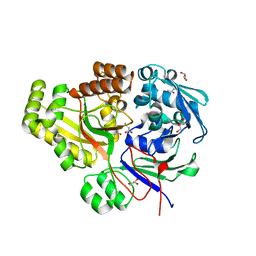 | | The Structural characterization of Burkholderia pseudomallei OppA. | | Descriptor: | CHLORIDE ION, GLYCEROL, OLIGOPEPTIDE DVA, ... | | Authors: | Lassaux, P, Gourlay, L.J, Bolognesi, M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Structure-Based Strategy for Epitope Discovery in Burkholderia Pseudomallei Oppa Antigen.
Structure, 21, 2013
|
|
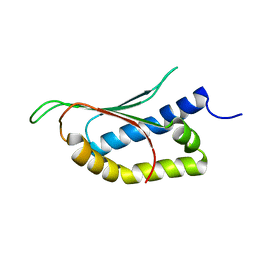
2MPE
 
 | |
