1Q9F
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX.
J.Mol.Biol., 336, 2004
|
|
1Q9G
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX
J.Mol.Biol., 336, 2004
|
|
1ORM
 
 | |
1CFE
 
 | | P14A, NMR, 20 STRUCTURES | | Descriptor: | PATHOGENESIS-RELATED PROTEIN P14A | | Authors: | Fernandez, C, Szyperski, T, Bruyere, T, Ramage, P, Mosinger, E, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the pathogenesis-related protein P14a.
J.Mol.Biol., 266, 1997
|
|
4ZJI
 
 | |
4ZJJ
 
 | |
4ZLO
 
 | | Serine/threonine-protein kinase PAK1 complexed with a dibenzodiazepine: identification of an allosteric site on PAK1 | | Descriptor: | 2,8-difluoro-11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine, GLYCEROL, Serine/threonine-protein kinase PAK 1 | | Authors: | Bellamacina, C.R, Bussiere, D.E. | | Deposit date: | 2015-05-01 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of a Dibenzodiazepine Hit to a Potent and Selective Allosteric PAK1 Inhibitor.
Acs Med.Chem.Lett., 6, 2015
|
|
5MVS
 
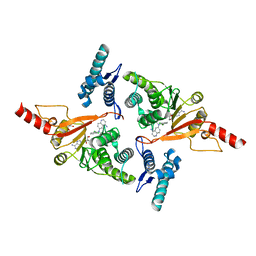 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | Descriptor: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW4
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD7 [N-(3-(((R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl)(methyl)amino)propyl)-2-(3-(2-chloro-3-(2-methylpyridin-3-yl)benzo[b]thiophen-5-yl)ureido)acetamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~2~-{[2-chloro-3-(2-methylpyridin-3-yl)-1-benzothiophen-5-yl]carbamoyl}-N-(3-{methyl[(3R)-1-(5H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]amino}propyl)glycinamide, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
8PD6
 
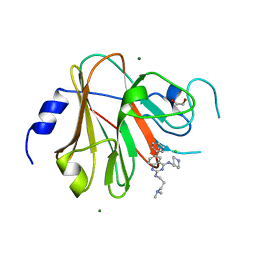 | | Crystal structure of the TRIM58 PRY-SPRY domain in complex with TRIM-473 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase TRIM58, ... | | Authors: | Renatus, M, Hoegenauer, K, Schroeder, M. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Ligands for TRIM58, a Novel Tissue-Selective E3 Ligase.
Acs Med.Chem.Lett., 14, 2023
|
|
8PD4
 
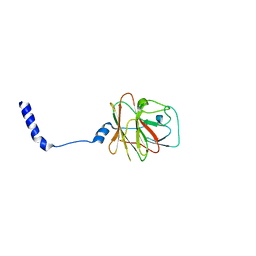 | |
5MW3
 
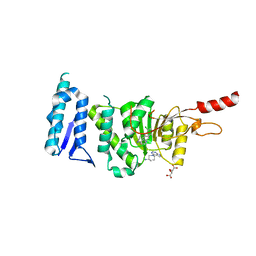 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | Descriptor: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
8OG6
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 1 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, ACETATE ION, DDB1- and CUL4-associated factor 1 | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGC
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-(2-azanylethylamino)-2-[1-(4-chlorophenyl)cyclopentyl]quinazolin-7-yl]piperazin-1-yl]ethanone, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG8
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-methyl-1-phenyl-propan-2-yl)imidazo[2,1-a]isoquinoline, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGA
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-(4-methoxyphenyl)cyclopropyl]-8-(4-methylpiperazin-1-yl)-2,3-dihydroimidazo[2,1-a]isoquinoline, DDB1- and CUL4-associated factor 1, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG5
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG7
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 2 | | Descriptor: | (4~{R})-4-[3-(4-chloranylphenoxy)phenyl]pyrrolidin-2-imine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG9
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 4 | | Descriptor: | 5-[1-(4-chlorophenyl)cyclopropyl]imidazo[2,1-a]isoquinoline, DDB1- and CUL4-associated factor 1 | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGB
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, DDB1- and CUL4-associated factor 1, DIMETHYL SULFOXIDE, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
5DTR
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD5 [N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5DTQ
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [(2,6-dichlorophenyl)(quinolin-6-yl)methanone] | | Descriptor: | (2,6-dichlorophenyl)(quinolin-6-yl)methanone, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5DTM
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide] | | Descriptor: | 4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
8A0V
 
 | | Crystal structure of TEAD3 in complex with CPD2 | | Descriptor: | DIMETHYL SULFOXIDE, MYRISTIC ACID, PHOSPHATE ION, ... | | Authors: | Scheufler, C, Kallen, J. | | Deposit date: | 2022-05-30 | | Release date: | 2022-08-17 | | Last modified: | 2022-10-19 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | The First Class of Small Molecules Potently Disrupting the YAP-TEAD Interaction by Direct Competition.
Chemmedchem, 17, 2022
|
|
8A0U
 
 | | Crystal structure of TEAD3 in complex with CPD4 | | Descriptor: | 2-[(2~{S})-2-(aminomethyl)-5-chloranyl-2-phenyl-3~{H}-1-benzofuran-4-yl]benzamide, MYRISTIC ACID, PHOSPHATE ION, ... | | Authors: | Scheufler, C, Kallen, J. | | Deposit date: | 2022-05-30 | | Release date: | 2022-08-17 | | Last modified: | 2022-10-19 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | The First Class of Small Molecules Potently Disrupting the YAP-TEAD Interaction by Direct Competition.
Chemmedchem, 17, 2022
|
|
