1NQL
 
 | |
1MAI
 
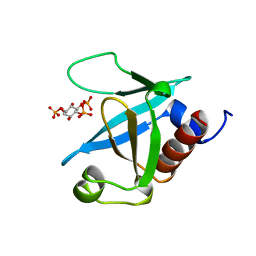 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM PHOSPHOLIPASE C DELTA IN COMPLEX WITH INOSITOL TRISPHOSPHATE | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOLIPASE C DELTA-1 | | Authors: | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | Deposit date: | 1996-05-23 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain.
Cell(Cambridge,Mass.), 83, 1995
|
|
1DYN
 
 | | CRYSTAL STRUCTURE AT 2.2 ANGSTROMS RESOLUTION OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM HUMAN DYNAMIN | | Descriptor: | DYNAMIN | | Authors: | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | Deposit date: | 1994-12-21 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin.
Cell(Cambridge,Mass.), 79, 1994
|
|
1FAO
 
 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM DAPP1/PHISH IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | DUAL ADAPTOR OF PHOSPHOTYROSINE AND 3-PHOSPHOINOSITIDES, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-07-13 | | Release date: | 2000-07-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
1FHX
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
1FB8
 
 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM DAPP1/PHISH | | Descriptor: | DUAL ADAPTOR OF PHOSPHOTYROSINE AND 3-PHOSPHOINOSITIDES, PHOSPHATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-07-14 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
1FHW
 
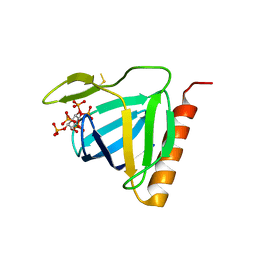 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol(1,3,4,5,6)pentakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
3B2U
 
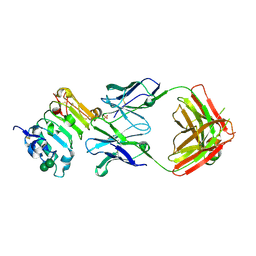 | | Crystal structure of isolated domain III of the extracellular region of the epidermal growth factor receptor in complex with the Fab fragment of IMC-11F8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Ferguson, K.M, Li, S, Kussie, P. | | Deposit date: | 2007-10-19 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis for EGF receptor inhibition by the therapeutic antibody IMC-11F8.
Structure, 16, 2008
|
|
3B2V
 
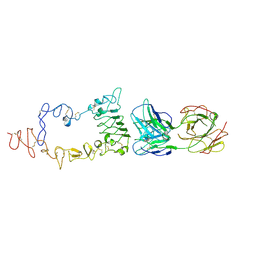 | | Crystal structure of the extracellular region of the epidermal growth factor receptor in complex with the Fab fragment of IMC-11F8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Ferguson, K.M, Li, S, Kussie, P. | | Deposit date: | 2007-10-19 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for EGF receptor inhibition by the therapeutic antibody IMC-11F8.
Structure, 16, 2008
|
|
4KRP
 
 | |
4KRL
 
 | |
4KRM
 
 | |
4KRO
 
 | |
4KRN
 
 | | Nanobody/VHH domain EgA1 | | Descriptor: | Nanobody/VHH domain EgA1, SULFATE ION | | Authors: | Ferguson, K.M, Schmitz, K.R. | | Deposit date: | 2013-05-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structural Evaluation of EGFR Inhibition Mechanisms for Nanobodies/VHH Domains.
Structure, 21, 2013
|
|
6B3S
 
 | |
3C09
 
 | | Crystal structure the Fab fragment of matuzumab (Fab72000) in complex with domain III of the extracellular region of EGFR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Matuzumab Fab Heavy chain, ... | | Authors: | Ferguson, K.M, Schmiedel, J, Knoechel, T. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization.
Cancer Cell, 13, 2008
|
|
3C08
 
 | |
3EGA
 
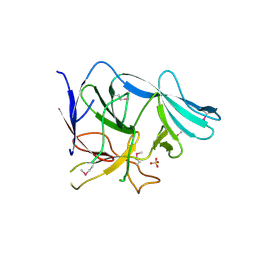 | |
3EGB
 
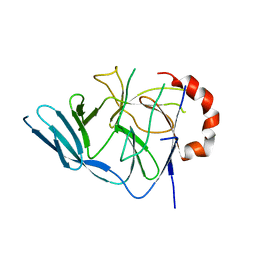 | |
2ZII
 
 | |
2ZIH
 
 | |
7LEN
 
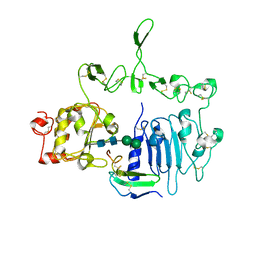 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with trehalose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LFR
 
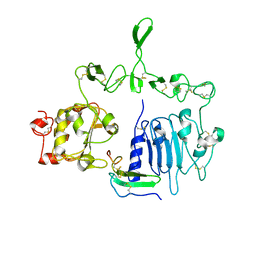 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with spermine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Proepiregulin, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LFS
 
 | | Crystal structure of the epidermal growth factor receptor extracellular region with A265V mutation in complex with epiregulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 4 of Epidermal growth factor receptor, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
3KN1
 
 | | Crystal Structure of Golgi Phosphoprotein 3 N-term Truncation Variant | | Descriptor: | Golgi phosphoprotein 3, SULFATE ION | | Authors: | Schmitz, K.R, Bessman, N.J, Setty, T.G, Ferguson, K.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking.
J.Cell Biol., 187, 2009
|
|
