1FI1
 
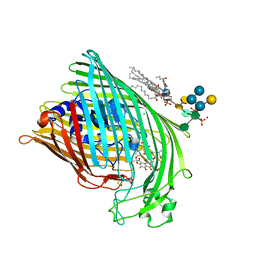 | | FhuA in complex with lipopolysaccharide and rifamycin CGP4832 | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Koedding, J, Boes, C, Walker, G, Coulton, J.W, Diederichs, K, Braun, V, Welte, W. | | Deposit date: | 2000-08-03 | | Release date: | 2001-08-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA.
Structure, 9, 2001
|
|
2Q7M
 
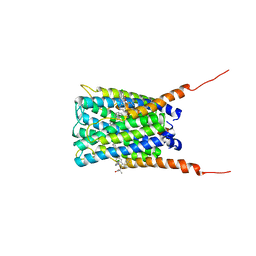 | | Crystal structure of human FLAP with MK-591 | | Descriptor: | 3-[3-(TERT-BUTYLTHIO)-1-(4-CHLOROBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
2Q7R
 
 | | Crystal structure of human FLAP with an iodinated analog of MK-591 | | Descriptor: | 3-[3-(3,3-DIMETHYLBUTANOYL)-1-(4-IODOBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
1QFG
 
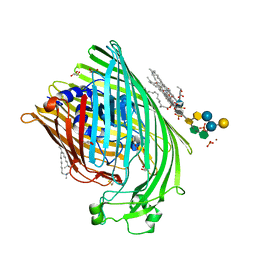 | | E. COLI FERRIC HYDROXAMATE RECEPTOR (FHUA) | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Welte, W, Hofmann, E, Lindner, B, Holst, O, Coulton, J.W, Diederichs, K. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
3S7F
 
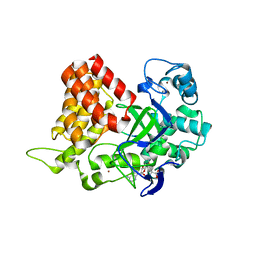 | |
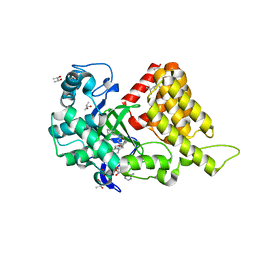
3S7D
 
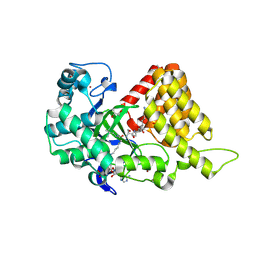 | |
3S7B
 
 | | Structural Basis of Substrate Methylation and Inhibition of SMYD2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[2-(5-hydroxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2011-05-26 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of Substrate Methylation and Inhibition of SMYD2.
Structure, 19, 2011
|
|
3S7J
 
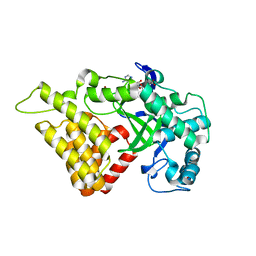 | |
3NGA
 
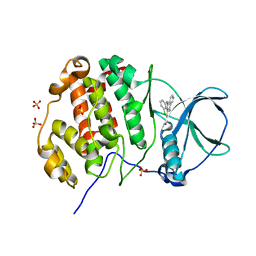 | | Human CK2 catalytic domain in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2010-06-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural basis of CX-4945 binding to human protein kinase CK2.
Febs Lett., 585, 2011
|
|
3NSZ
 
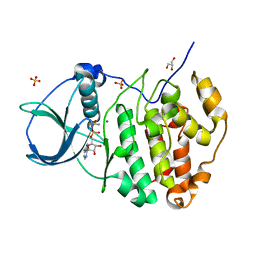 | | Human CK2 catalytic domain in complex with AMPPN | | Descriptor: | Casein kinase II subunit alpha, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of CX-4945 binding to human protein kinase CK2.
Febs Lett., 585, 2011
|
|
1QJQ
 
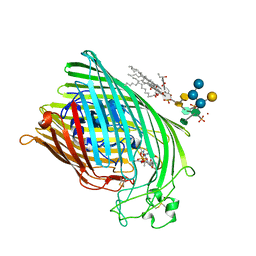 | | FERRIC HYDROXAMATE RECEPTOR FROM ESCHERICHIA COLI (FHUA) | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DIPHOSPHATE, FERRIC HYDROXAMATE RECEPTOR, ... | | Authors: | Ferguson, A.D, Braun, V, Fiedler, H.-P, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-06-29 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA.
Protein Sci., 9, 2000
|
|
1QFF
 
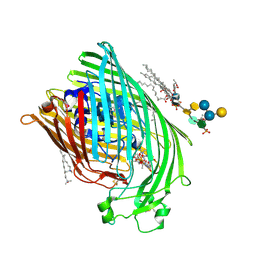 | | E. COLI FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) IN COMPLEX WITH BOUND FERRICHROME-IRON | | Descriptor: | 2-AMINO-VINYL-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Ferguson, A.D, Hofmann, E, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
1QKC
 
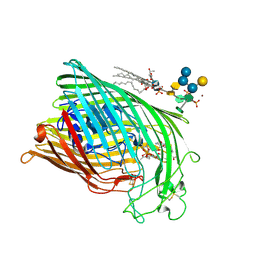 | | ESCHERICHIA COLI FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) IN COMPLEX DELTA TWO-ALBOMYCIN | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DELTA-2-ALBOMYCIN A1, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Braun, V, Fiedler, H.-P, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-07-18 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA.
Protein Sci., 9, 2000
|
|
5KGG
 
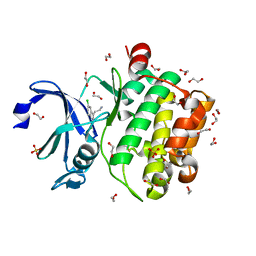 | |
5KGK
 
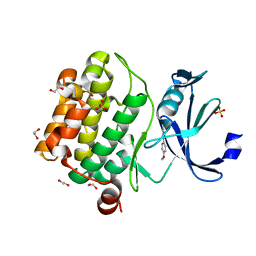 | |
1ZZV
 
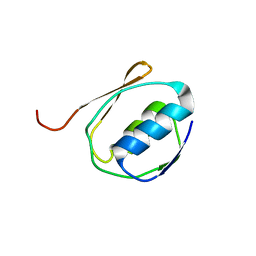 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter FecA from Escherichia coli. | | Descriptor: | Iron(III) dicitrate transport protein fecA | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-14 | | Release date: | 2006-09-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
2A02
 
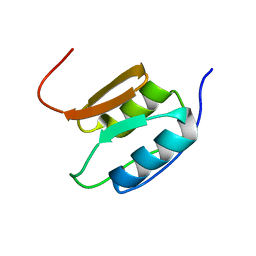 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter PupA from Pseudomonas putida. | | Descriptor: | Ferric-pseudobactin 358 receptor | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-15 | | Release date: | 2006-09-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
2A4H
 
 | | Solution structure of Sep15 from Drosophila melanogaster | | Descriptor: | Selenoprotein Sep15 | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
2A2P
 
 | | Solution structure of SelM from Mus musculus | | Descriptor: | Selenoprotein M | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
5KGE
 
 | |
5KGI
 
 | |
5KGD
 
 | |
5KWH
 
 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CK2
To Be Published
|
|
5KU8
 
 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D, Dowling, J. | | Deposit date: | 2016-07-13 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of CK2
Not Published
|
|
1KMP
 
 | | Crystal structure of the Outer Membrane Transporter FecA Complexed with Ferric Citrate | | Descriptor: | CITRIC ACID, FE (III) ION, IRON(III) DICITRATE TRANSPORT PROTEIN FECA, ... | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
