4PX7
 
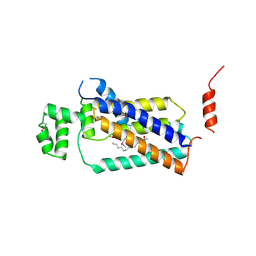 | | Crystal structure of lipid phosphatase E. coli PgpB | | Descriptor: | GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, Phosphatidylglycerophosphatase | | Authors: | Fan, J, Jiang, D, Zhao, Y, Zhang, X.C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-05-28 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6V8Q
 
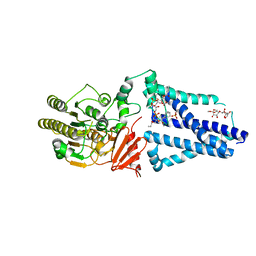 | | Structure of an inner membrane protein required for PhoPQ regulated increases in outer membrane cardiolipin | | Descriptor: | (9Z,21R,24R,30R,33R,44Z)-24,27,30-trihydroxy-18,24,30,36-tetraoxo-19,23,25,29,31,35-hexaoxa-24lambda~5~,30lambda~5~-dip hosphatripentaconta-9,44-diene-21,33-diyl (9Z,9'Z)di-octadec-9-enoate, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fan, J, Miller, S. | | Deposit date: | 2019-12-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.699585 Å) | | Cite: | Structure of an Inner Membrane Protein Required for PhoPQ-Regulated Increases in Outer Membrane Cardiolipin.
Mbio, 11, 2020
|
|
2INF
 
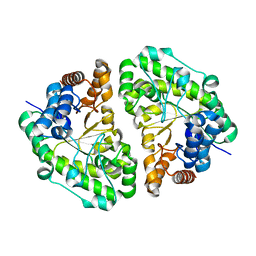 | | Crystal Structure of Uroporphyrinogen Decarboxylase from Bacillus subtilis | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Fan, J, Liu, Q, Hao, Q, Teng, M.K, Niu, L.W. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uroporphyrinogen decarboxylase from Bacillus subtilis
J.Bacteriol., 189, 2007
|
|
7DJC
 
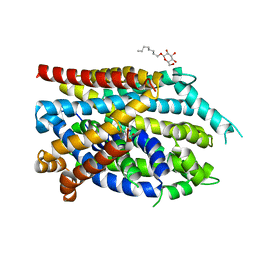 | | Crystal structure of the G26C/Q250A mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ1
 
 | | Crystal structure of the G26C mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.528 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DII
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 7 | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DIX
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 5 | | Descriptor: | Na(+):neurotransmitter symporter (Snf family), SELENOMETHIONINE, SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ2
 
 | | Crystal structure of the G26C/E290S mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7XJ0
 
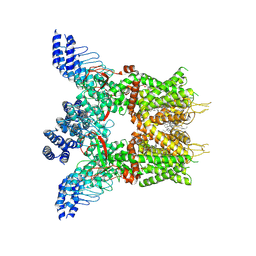 | | Structure of human TRPV3 in complex with Trpvicin | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ3
 
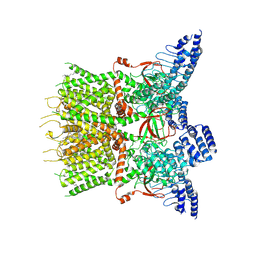 | | Structure of human TRPV3 | | Descriptor: | [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, fusion of transient receptor potential cation channel subfamily V member 3 and 3C-GFP | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ2
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C4 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ1
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
8WR2
 
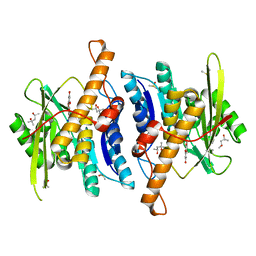 | | Crystal Structure of Human Pyridoxal Kinase with bound Luteolin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Fan, J, Zhu, Y. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and characterization of natural product luteolin as an effective inhibitor of human pyridoxal kinase.
Bioorg.Chem., 143, 2024
|
|
4CKH
 
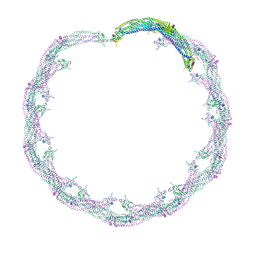 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
4CKG
 
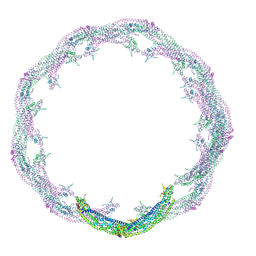 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
8SQF
 
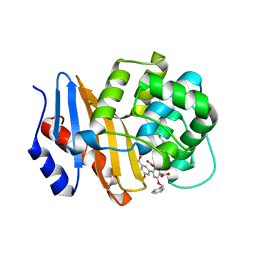 | | OXA-48 bound to inhibitor CDD-2725 | | Descriptor: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8SQG
 
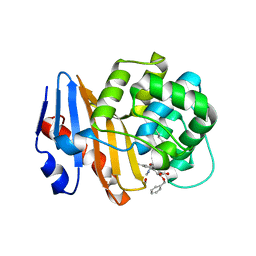 | | OXA-48 bound to inhibitor CDD-2801 | | Descriptor: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
6QJP
 
 | | Cryo-EM structure of heparin-induced 2N4R tau jagged filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJH
 
 | | Cryo-EM structure of heparin-induced 2N4R tau snake filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJM
 
 | | Cryo-EM structure of heparin-induced 2N4R tau twister filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJQ
 
 | | Cryo-EM structure of heparin-induced 2N3R tau filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
3BS8
 
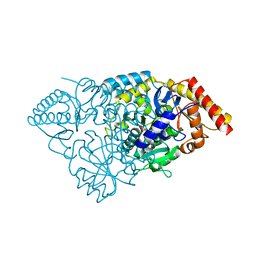 | | Crystal structure of Glutamate 1-Semialdehyde Aminotransferase complexed with pyridoxamine-5'-phosphate From Bacillus subtilis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Ge, H, Fan, J, Teng, M, Niu, L. | | Deposit date: | 2007-12-22 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Glutamate1-semialdehyde aminotransferase from Bacillus subtilis with bound pyridoxamine-5'-phosphate
Biochem.Biophys.Res.Commun., 402, 2010
|
|
5H3D
 
 | | Helical structure of membrane tubules decorated by ACAP1 (BARPH doamin) protein by cryo-electron microscopy and MD simulation | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Chan, C, Pang, X.Y, Zhang, Y, Sun, F, Fan, J. | | Deposit date: | 2016-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | ACAP1 assembles into an unusual protein lattice for membrane deformation through multiple stages.
Plos Comput.Biol., 15, 2019
|
|
5JQM
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 1.5 Angstroms Resolution | | Descriptor: | Protein UPS1, mitochondrial,Mitochondrial distribution and morphology protein 35 | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
5JQL
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 2.9 Angstroms Resolution | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
