1EZ3
 
 | |
1KHX
 
 | | Crystal structure of a phosphorylated Smad2 | | Descriptor: | Smad2 | | Authors: | Wu, J.-W, Hu, M, Chai, J, Seoane, J, Huse, M, Kyin, S, Muir, T.W, Fairman, R, Massague, J, Shi, Y. | | Deposit date: | 2001-12-01 | | Release date: | 2002-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling.
Mol.Cell, 8, 2001
|
|
1NW9
 
 | | STRUCTURE OF CASPASE-9 IN AN INHIBITORY COMPLEX WITH XIAP-BIR3 | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ZINC ION, caspase 9, ... | | Authors: | Shiozaki, E.N, Chai, J, Rigotti, D.J, Riedl, S.J, Li, P, Srinivasula, S.M, Alnemri, E.S, Fairman, R, Shi, Y. | | Deposit date: | 2003-02-05 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of XIAP-Mediated Inhibition of Caspase-9
Mol.Cell, 11, 2003
|
|
1PV8
 
 | | Crystal structure of a low activity F12L mutant of human porphobilinogen synthase | | Descriptor: | 3-(2-AMINOETHYL)-4-(AMINOMETHYL)HEPTANEDIOIC ACID, Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Breinig, S, Kervinen, J, Stith, L, Wasson, A.S, Fairman, R, Wlodawer, A, Zdanov, A, Jaffe, E.K. | | Deposit date: | 2003-06-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase.
Nat.Struct.Biol., 10, 2003
|
|
2AR9
 
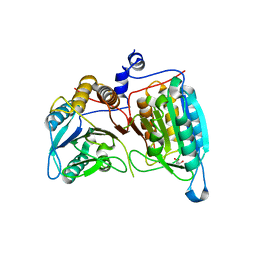 | | Crystal structure of a dimeric caspase-9 | | Descriptor: | Caspase-9, D-MALATE | | Authors: | Chao, Y, Shiozaki, E.N, Srinivassula, S.M, Rigotti, D.J, Fairman, R, Shi, Y. | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Engineering a Dimeric Caspase-9: A Re-Evaluation of the Induced Proximity Model for Caspase Activation
PLOS BIOL., 3, 2005
|
|
6U3W
 
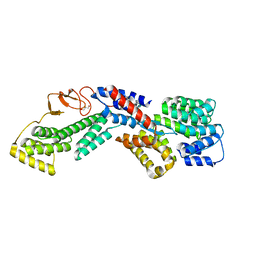 | |
2F3O
 
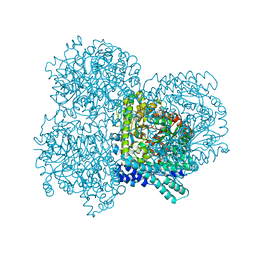 | |
6U3V
 
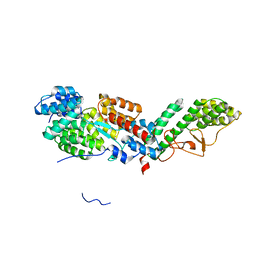 | |
6TZT
 
 | |
1TW6
 
 | | Structure of an ML-IAP/XIAP chimera bound to a 9mer peptide derived from Smac | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Vucic, D, Wallweber, H.J.A, Das, K, Shin, H, Elliott, L.O, Kadkhodayan, S, Deshayes, K, Salvesen, G.S, Fairbrother, W.J. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.713 Å) | | Cite: | Engineering ML-IAP to produce an extraordinarily potent caspase 9 inhibitor: implications for Smac-dependent anti-apoptotic activity of ML-IAP
Biochem.J., 385, 2005
|
|
5EP0
 
 | | Quorum-Sensing Signal Integrator LuxO - Receiver+Catalytic Domains | | Descriptor: | 1,2-ETHANEDIOL, Putative repressor protein luxO, SULFATE ION | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP2
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain in Complex with AzaU Inhibitor | | Descriptor: | 2,2-dimethylpropyl 2-[[3,5-bis(oxidanylidene)-2~{H}-1,2,4-triazin-6-yl]sulfanyl]ethanoate, ACETATE ION, Putative repressor protein luxO | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP1
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain | | Descriptor: | ACETATE ION, Putative repressor protein luxO | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP4
 
 | | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EP3
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain Bound to CV-133 Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,2-dimethylpropyl 2-[(3-oxidanylidene-5-sulfanylidene-2~{H}-1,2,4-triazin-6-yl)amino]ethanoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
