4XJX
 
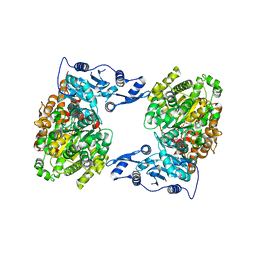 | | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HsdR, MAGNESIUM ION | | Authors: | Baikova, T, Stsiapanava, A, Moche, M, Degtjarik, O, Kuta-Smatanova, I, Ettrich, R. | | Deposit date: | 2015-01-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP
To Be Published
|
|
3ZHO
 
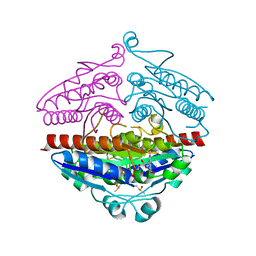 | | X-ray structure of E.coli Wrba in complex with FMN at 1.2 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOPROTEIN WRBA | | Authors: | Kishko, I, Lapkouski, M, Brynda, J, Kuty, M, Carey, J, Smatanova, I.K, Ettrich, R. | | Deposit date: | 2012-12-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 A Resolution Crystal Structure of Escherichia Coli Wrba Holoprotein
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2VU4
 
 | | Structure of PsbP protein from Spinacia oleracea at 1.98 A resolution | | Descriptor: | OXYGEN-EVOLVING ENHANCER PROTEIN 2, ZINC ION | | Authors: | Lapkouski, M, Ristvejova, R, Arellano, J.B, Revuelta, J.L, Kuta Smatanova, I, Ettrich, R. | | Deposit date: | 2008-05-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Raman Spectroscopy Adds Complementary Detail to the High-Resolution X-Ray Crystal Structure of Photosynthetic Psbp from Spinacia Oleracea.
Plos One, 7, 2012
|
|
5F12
 
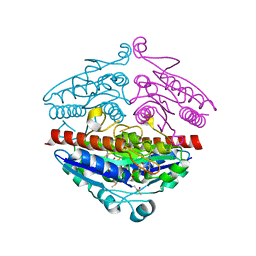 | | WrbA in complex with FMN under crystallization conditions of WrbA-FMN-BQ structure (4YQE) | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone) | | Authors: | Degtjarik, O, Brynda, J, Ettrichova, O, Kuta Smatanova, I, Carey, J, Ettrich, R. | | Deposit date: | 2015-11-29 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Quantum Calculations Indicate Effective Electron Transfer between FMN and Benzoquinone in a New Crystal Structure of Escherichia coli WrbA.
J.Phys.Chem.B, 120, 2016
|
|
4YQE
 
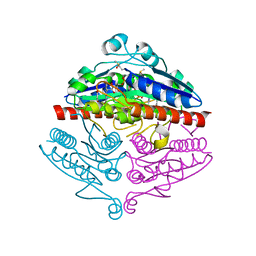 | | Crystal structure of E. coli WrbA in complex with benzoquinone | | Descriptor: | 1,4-benzoquinone, FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone) | | Authors: | Degtjarik, O, Brynda, J, Ettrichova, O, Carey, J, Kuta Smatanova, I, Ettrich, R. | | Deposit date: | 2015-03-13 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Quantum Calculations Indicate Effective Electron Transfer between FMN and Benzoquinone in a New Crystal Structure of Escherichia coli WrbA.
J.Phys.Chem.B, 120, 2016
|
|
2W00
 
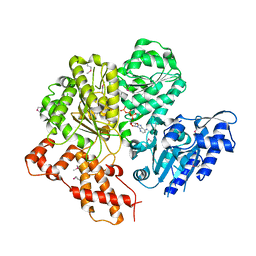 | | Crystal structure of the HsdR subunit of the EcoR124I restriction enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HSDR, MAGNESIUM ION | | Authors: | Lapkouski, M, Panjikar, S, Kuta Smatanova, I, Ettrich, R, Csefalvay, E. | | Deposit date: | 2008-08-08 | | Release date: | 2008-12-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Motor Subunit of Type I Restriction-Modification Complex Ecor124I.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4DY4
 
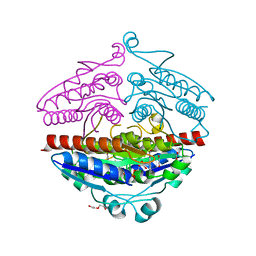 | | High resolution structure of E.coli WrbA with FMN | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, Flavoprotein wrbA | | Authors: | Kishko, I, Brynda, J, Kuta Smatanova, I, Ettrich, R, Carey, J. | | Deposit date: | 2012-02-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution structure of E.coli WrbA with FMN
To be Published
|
|
4BEC
 
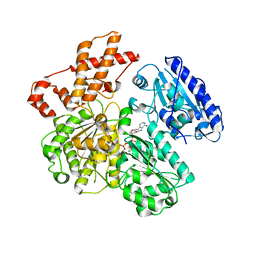 | | MUTANT (K220A) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4BE7
 
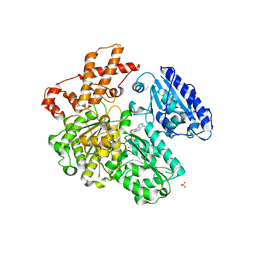 | | MUTANT (K220R) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4BEB
 
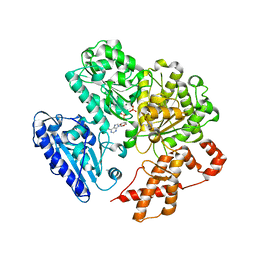 | | MUTANT (K220E) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
5J3N
 
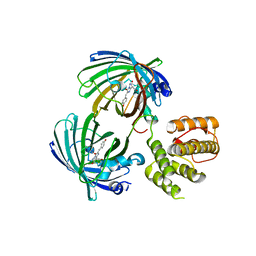 | | C-terminal domain of EcoR124I HsdR subunit fused with the pH-sensitive GFP variant ratiometric pHluorin | | Descriptor: | Green fluorescent protein,HsdR | | Authors: | Grinkevich, P, Iermak, I, Luedtke, N, Mesters, J.R, Ettrich, R, Ludwig, J. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a novel domain of the motor subunit of the Type I restriction enzyme EcoR124 involved in complex assembly and DNA binding.
J. Biol. Chem., 293, 2018
|
|
2R96
 
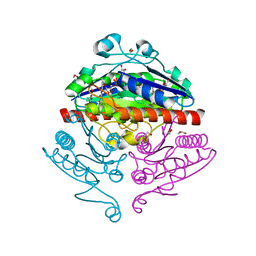 | | Crystal structure of E. coli WrbA in complex with FMN | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
2R97
 
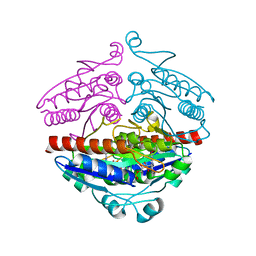 | | Crystal structure of E. coli WrbA in complex with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
2MWQ
 
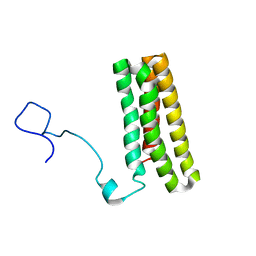 | | Solution structure of PsbQ from spinacia oleracea | | Descriptor: | Oxygen-evolving enhancer protein 3, chloroplastic | | Authors: | Rathner, P, Mueller, N, Wimmer, R, Chandra, K. | | Deposit date: | 2014-11-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR and molecular dynamics reveal a persistent alpha helix within the dynamic region of PsbQ from photosystem II of higher plants.
Proteins, 83, 2015
|
|
2RG1
 
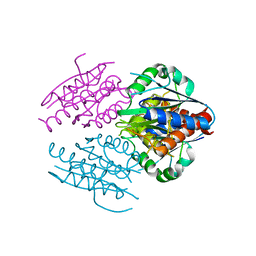 | | Crystal structure of E. coli WrbA apoprotein | | Descriptor: | CHLORIDE ION, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Lapkouski, M, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
4E46
 
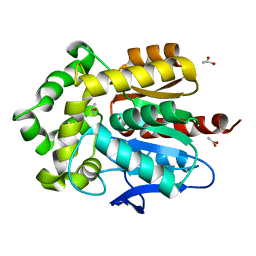 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA in complex with 2-propanol | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Stsiapanava, A, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Expansion of access tunnels and active-site cavities influence activity of haloalkane dehalogenases in organic cosolvents.
Chembiochem, 14, 2013
|
|
4F60
 
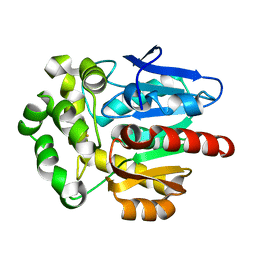 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (T148L, G171Q, A172V, C176F). | | Descriptor: | FLUORIDE ION, Haloalkane dehalogenase | | Authors: | Plevaka, M, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4F5Z
 
 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (L95V, A172V). | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Kulik, D, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
