1BH6
 
 | | SUBTILISIN DY IN COMPLEX WITH THE SYNTHETIC INHIBITOR N-BENZYLOXYCARBONYL-ALA-PRO-PHE-CHLOROMETHYL KETONE | | Descriptor: | CALCIUM ION, N-BENZYLOXYCARBONYL-ALA-PRO-3-AMINO-4-PHENYL-BUTAN-2-OL, SODIUM ION, ... | | Authors: | Eschenburg, S, Genov, N, Wilson, K.S, Betzel, C. | | Deposit date: | 1998-06-15 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of subtilisin DY, a random mutant of subtilisin Carlsberg.
Eur.J.Biochem., 257, 1998
|
|
1EJD
 
 | | Crystal structure of unliganded mura (type1) | | Descriptor: | CYCLOHEXYLAMMONIUM ION, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
1EJC
 
 | | Crystal structure of unliganded mura (type2) | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
1YBG
 
 | | MurA inhibited by a derivative of 5-sulfonoxy-anthranilic acid | | Descriptor: | N-METHYL-N-{2-[(2-NAPHTHYLSULFONYL)AMINO]-5-[(2-NAPHTHYLSULFONYL)OXY]BENZOYL}-L-ASPARTIC ACID, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Priestman, M.A, Abdul-Latif, F.A, Delachaume, C, Fassy, F, Schonbrunn, E. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Inhibitor That Suspends the Induced Fit Mechanism of UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA).
J.Biol.Chem., 280, 2005
|
|
1Q36
 
 | | EPSP synthase (Asp313Ala) liganded with tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, 5-(1-CARBOXY-1-PHOSPHONOOXY-ETHOXYL)-4-HYDROXY-3-PHOSPHONOOXY-CYCLOHEX-1-ENECARBOXYLIC ACID, FORMIC ACID | | Authors: | Eschenburg, S, Kabsch, W, Healy, M.L, Schonbrunn, E. | | Deposit date: | 2003-07-28 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A New View of the Mechanisms of UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA) and 5-Enolpyruvylshikimate-3-phosphate Synthase (AroA) Derived from X-ray Structures of Their Tetrahedral Reaction Intermediate States.
J.Biol.Chem., 278, 2003
|
|
1Q3G
 
 | | MurA (Asp305Ala) liganded with tetrahedral reaction intermediate | | Descriptor: | 1,2-ETHANEDIOL, 3'-1-CARBOXY-1-PHOSPHONOOXY-ETHOXY-URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Kabsch, W, Healy, M.L, Schonbrunn, E. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A New View of the Mechanisms of UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA) and 5-Enolpyruvylshikimate-3-phosphate Synthase (AroA) Derived from X-ray Structures of Their Tetrahedral Reaction Intermediate States.
J.Biol.Chem., 278, 2003
|
|
1RYW
 
 | | C115S MurA liganded with reaction products | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2003-12-22 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence That the Fosfomycin Target Cys115 in UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA) Is Essential for Product Release.
J.Biol.Chem., 280, 2005
|
|
1MI4
 
 | | Glyphosate insensitive G96A mutant EPSP synthase liganded with shikimate-3-phosphate | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Eschenburg, S, Healy, M.L, Priestman, M.A, Lushington, G.H, Schonbrunn, E. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | How the mutation glycine96 to alanine confers glyphosate insensitivity to 5-enolpyruvyl
shikimate-3-phosphate synthase from Escherichia coli.
PLANTA, 216, 2002
|
|
3SFZ
 
 | | Crystal structure of full-length murine Apaf-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apoptotic peptidase activating factor 1, GAMMA-BUTYROLACTONE | | Authors: | Eschenburg, S, Reubold, T.F. | | Deposit date: | 2011-06-14 | | Release date: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of full-length Apaf-1: how the death signal is relayed in the mitochondrial pathway of apoptosis.
Structure, 19, 2011
|
|
3SHF
 
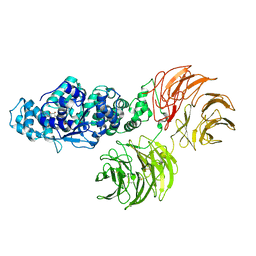 | |
1AOK
 
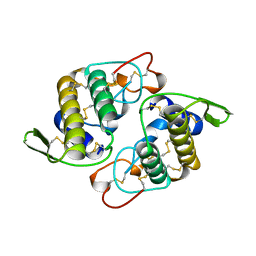 | | VIPOXIN COMPLEX | | Descriptor: | ACETATE ION, VIPOXIN COMPLEX | | Authors: | Perbandt, M, Wilson, J.C, Eschenburg, S, Betzel, C. | | Deposit date: | 1997-07-07 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of vipoxin at 2.0 A: an example of regulation of a toxic function generated by molecular evolution.
FEBS Lett., 412, 1997
|
|
8QDO
 
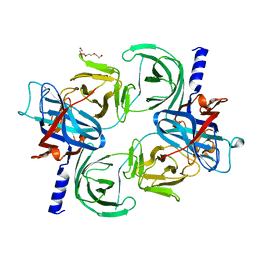 | | Crystal structure of the tegument protein UL82 (pp71) from Human Cytomegalovirus | | Descriptor: | Protein pp71, TETRAETHYLENE GLYCOL | | Authors: | Bresch, I.P, Eberhage, J, Reubold, T.F, Eschenburg, S. | | Deposit date: | 2023-08-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the tegument protein UL82 (pp71) from human cytomegalovirus.
Protein Sci., 33, 2024
|
|
2AKA
 
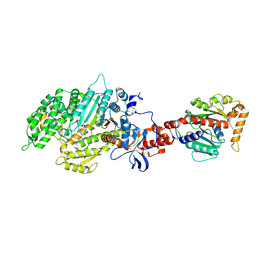 | | Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin 1 from Rattus norvegicus | | Descriptor: | Dynamin-1, LINKER, myosin II heavy chain | | Authors: | Reubold, T.F, Eschenburg, S, Becker, A, Leonard, M, Schmid, S.L, Vallee, R.B, Kull, F.J, Manstein, D.J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GTPase domain of rat dynamin 1.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2MLL
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-16 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
7ZGU
 
 | | Human NLRP3-deltaPYD hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Raisch, T, Machtens, D.A, Bresch, I.B, Eberhage, J, Prumbaum, D, Reubold, T.F, Raunser, S, Eschenburg, S. | | Deposit date: | 2022-04-04 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the NEK7-independent NLRP3 inflammasome
To Be Published
|
|
6YVG
 
 | | Crystal structure of MesI (Lpg2505) from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MesI (Lpg2505) | | Authors: | Machtens, D.A, Willerding, J.M, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the metaeffector MesI (Lpg2505) from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
4IM6
 
 | | LRR domain from human NLRP1 | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Hahne, G, Reubold, T.F, Eschenburg, S. | | Deposit date: | 2013-01-02 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the leucine-rich repeat domain of the NOD-like receptor NLRP1: implications for binding of muramyl dipeptide.
Febs Lett., 588, 2014
|
|
8BVP
 
 | | Crystal structure of an N-terminal fragment of the effector protein Lpg2504 (SidI) from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Restriction endonuclease | | Authors: | Machtens, D.A, Willerding, J.M, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the N-terminal domain of the effector protein SidI of Legionella pneumophila reveals a glucosyl transferase domain.
Biochem.Biophys.Res.Commun., 661, 2023
|
|
5A3F
 
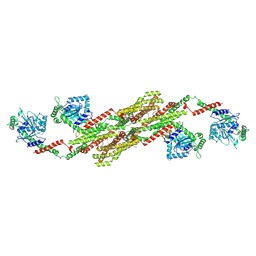 | | Crystal structure of the dynamin tetramer | | Descriptor: | DYNAMIN 3 | | Authors: | Reubold, T.F, Faelber, K, Plattner, N, Posor, Y, Branz, K, Curth, U, Schlegel, J, Anand, R, Manstein, D.J, Noe, F, Haucke, V, Daumke, O, Eschenburg, S. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the Dynamin Tetramer
Nature, 525, 2015
|
|
5D3Q
 
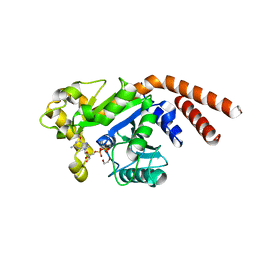 | | Dynamin 1 GTPase-BSE fusion dimer complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Dynamin-1,Dynamin-1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Anand, R, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the GTPase domain and the bundle signalling element of dynamin in the GDP state.
Biochem.Biophys.Res.Commun., 469, 2016
|
|
1EYN
 
 | | Structure of mura liganded with the extrinsic fluorescence probe ANS | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, GLYCEROL, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYLTRANSFERASE | | Authors: | Schonbrunn, E, Eschenburg, S, Luger, K, Kabsch, W, Amrhein, N. | | Deposit date: | 2000-05-07 | | Release date: | 2000-06-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1NAW
 
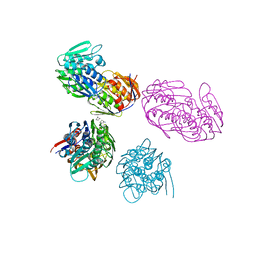 | | ENOLPYRUVYL TRANSFERASE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | Authors: | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | Deposit date: | 1996-07-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
1IC6
 
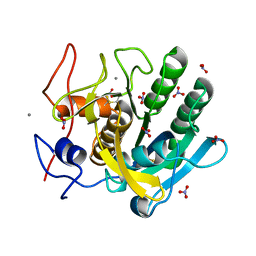 | | STRUCTURE OF A SERINE PROTEASE PROTEINASE K FROM TRITIRACHIUM ALBUM LIMBER AT 0.98 A RESOLUTION | | Descriptor: | CALCIUM ION, NITRATE ION, PROTEINASE K | | Authors: | Betzel, C, Gourinath, S, Kumar, P, Kaur, P, Perbandt, M, Eschenburg, S, Singh, T.P. | | Deposit date: | 2001-03-30 | | Release date: | 2001-04-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure of a serine protease proteinase K from Tritirachium album limber at 0.98 A resolution.
Biochemistry, 40, 2001
|
|
1DLG
 
 | | CRYSTAL STRUCTURE OF THE C115S ENTEROBACTER CLOACAE MURA IN THE UN-LIGANDED STATE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE MURA | | Authors: | Schonbrunn, E, Eschenburg, S, Krekel, F, Luger, K, Amrhein, N. | | Deposit date: | 1999-12-09 | | Release date: | 2000-04-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the loop containing residue 115 in the induced-fit mechanism of the bacterial cell wall biosynthetic enzyme MurA.
Biochemistry, 39, 2000
|
|
1CE7
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-18 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
