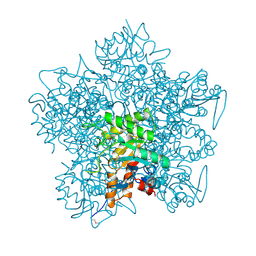
1AW5
 
 | |
1B4E
 
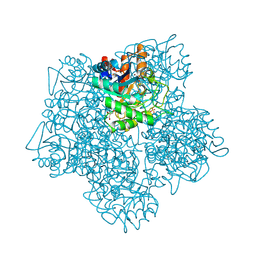 | | X-ray structure of 5-aminolevulinic acid dehydratase complexed with the inhibitor levulinic acid | | Descriptor: | GLYCEROL, LAEVULINIC ACID, PROTEIN (5-AMINOLEVULINIC ACID DEHYDRATASE), ... | | Authors: | Erskine, P.T, Cooper, J.B, Lewis, G, Spencer, P, Wood, S.P, Shoolingin-Jordan, P.M. | | Deposit date: | 1998-12-19 | | Release date: | 1999-12-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 5-aminolevulinic acid dehydratase from Escherichia coli complexed with the inhibitor levulinic acid at 2.0 A resolution.
Biochemistry, 38, 1999
|
|
1H7P
 
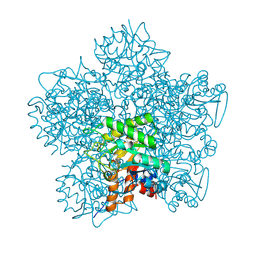 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 4-KETO-5-AMINO-HEXANOIC (KAH) AT 1.64 A RESOLUTION | | Descriptor: | 5-AMINO-4-HYDROXYHEXANOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1H7O
 
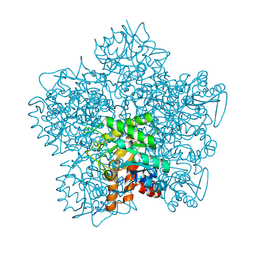 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 5-AMINOLAEVULINIC ACID AT 1.7 A RESOLUTION | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, DELTA-AMINO VALERIC ACID, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1H7N
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH LAEVULINIC ACID AT 1.6 A RESOLUTION | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1H7R
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH SUCCINYLACETONE AT 2.0 A RESOLUTION. | | Descriptor: | 4,6-DIOXOHEPTANOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1YLV
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH LAEVULINIC ACID | | Descriptor: | LAEVULINIC ACID, PROTEIN (5-AMINOLAEVULINIC ACID DEHYDRATASE), ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Roper, J, Coker, A, Warren, M.J, Shoolingin-Jordan, P.M, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-02-22 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Schiff base complex of yeast 5-aminolaevulinic acid dehydratase with laevulinic acid.
Protein Sci., 8, 1999
|
|
1W31
 
 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 5-HYDROXYLAEVULINIC ACID COMPLEX | | Descriptor: | 5-HYDROXYLAEVULINIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Beaven, G.D.E, Gill, R, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2004-07-11 | | Release date: | 2005-08-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with the Inhibitor 5-Hydroxylaevulinic Acid
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2CCM
 
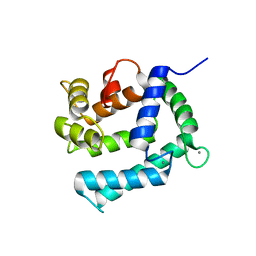 | | X-ray structure of Calexcitin from Loligo pealeii at 1.8A | | Descriptor: | CALCIUM ION, CALEXCITIN | | Authors: | Erskine, P.T, Beaven, G.D.E, Wood, S.P, Fox, G, Vernon, J, Giese, K.P, Cooper, J.B. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Neuronal Protein Calexcitin Suggests a Mode of Interaction in Signalling Pathways of Learning and Memory.
J.Mol.Biol., 357, 2006
|
|
2IZP
 
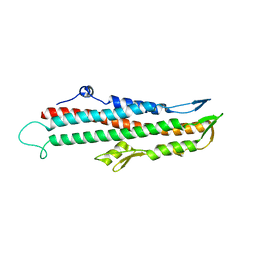 | | BipD - an invasion protein associated with the type-III secretion system of Burkholderia pseudomallei. | | Descriptor: | PUTATIVE MEMBRANE ANTIGEN | | Authors: | Erskine, P.T, Knight, M.J, Ruaux, A, Mikolajek, H, Wong-Fat-Sang, N, Withers, J, Gill, R, Wood, S.P, Wood, M, Fox, G.C, Cooper, J.B. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-06 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution Structure of Bipd: An Invasion Protein Associated with the Type III Secretion System of Burkholderia Pseudomallei.
J.Mol.Biol., 363, 2006
|
|
1GJP
 
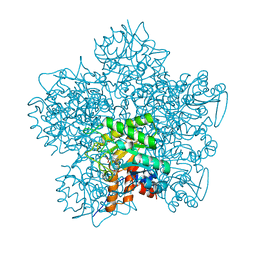 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 4-OXOSEBACIC ACID | | Descriptor: | 4-OXODECANEDIOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1OHL
 
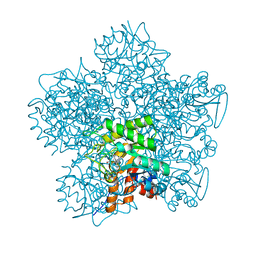 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE PUTATIVE CYCLIC REACTION INTERMEDIATE COMPLEX | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, BETA-MERCAPTOETHANOL, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ... | | Authors: | Erskine, P.T, Coates, L, Butler, D, Youell, J.H, Brindley, A.A, Wood, S.P, Warren, M.J, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Structure of a Putative Reaction Intermediateof 5-Aminolaevulinic Acid Dehydratase
Biochem.J., 373, 2003
|
|
1QML
 
 | | Hg complex of yeast 5-aminolaevulinic acid dehydratase | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, MERCURY (II) ION | | Authors: | Erskine, P.T, Senior, N, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-02 | | Release date: | 2000-10-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNV
 
 | | yeast 5-aminolaevulinic acid dehydratase Lead (Pb) complex | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LEAD (II) ION | | Authors: | Erskine, P.T, Senior, N.M, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 2000-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4NDB
 
 | | X-ray structure of a mutant (T61D) of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NDC
 
 | | X-ray structure of a mutant (T188D) of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NDD
 
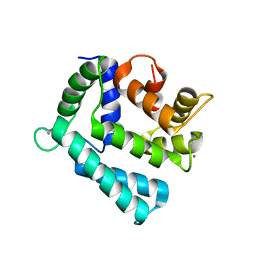 | | X-ray structure of a double mutant of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1EB3
 
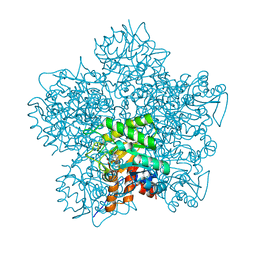 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 4,7-DIOXOSEBACIC ACID COMPLEX | | Descriptor: | 4,7-DIOXOSEBACIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-07-18 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1OEW
 
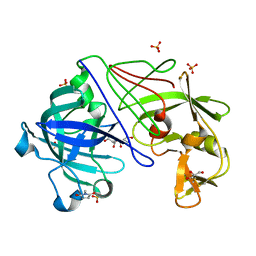 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OEX
 
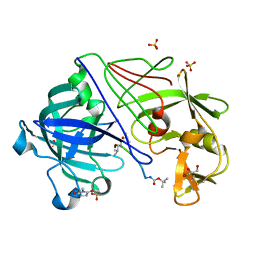 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
3NFT
 
 | | Near-atomic resolution analysis of BipD- A component of the type-III secretion system of Burkholderia pseudomallei | | Descriptor: | Translocator protein bipD | | Authors: | Pal, M, Erskine, P.T, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Near-atomic resolution analysis of BipD, a component of the type III secretion system of Burkholderia pseudomallei.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4HTG
 
 | | Porphobilinogen Deaminase from Arabidopsis Thaliana | | Descriptor: | 3-[(5Z)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methylidene}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, ACETATE ION, Porphobilinogen deaminase, ... | | Authors: | Roberts, A, Gill, R, Hussey, R.J, Erskine, P.T, Cooper, J.B, Wood, S.P, Chrystal, E.J.T, Shoolingin-Jordan, P.M. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanism of pyrrole polymerization catalysed by porphobilinogen deaminase: high-resolution X-ray studies of the Arabidopsis thaliana enzyme.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1W6S
 
 | | The high resolution structure of methanol dehydrogenase from methylobacterium extorquens | | Descriptor: | CALCIUM ION, GLYCEROL, METHANOL DEHYDROGENASE SUBUNIT 1, ... | | Authors: | Williams, P.A, Coates, L, Mohammed, F, Gill, R, Erskine, P.T, Wood, S.P, Anthony, C, Cooper, J.B. | | Deposit date: | 2004-08-23 | | Release date: | 2004-12-21 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Atomic Resolution Structure of Methanol Dehydrogenase from Methylobacterium Extorquens
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W1Z
 
 | | Structure of the plant like 5-Aminolaevulinic Acid Dehydratase from Chlorobium vibrioforme | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S.I, Avissar, Y.J, Gill, R, Mohammed, F, Wood, S.P, Shoolingin-Jordan, P, Cooper, J.B. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the plant like 5-aminolaevulinic acid dehydratase from Chlorobium vibrioforme complexed with the inhibitor laevulinic acid at 2.6 A resolution.
J. Mol. Biol., 342, 2004
|
|
5A92
 
 | | 15K X-ray structure with Cefotaxime: Exploring the Mechanism of beta- Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | Descriptor: | BETA-LACTAMASE CTX-M-97, CEFOTAXIME, C3' cleaved, ... | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
