1N7S
 
 | | High Resolution Structure of a Truncated Neuronal SNARE Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, SNAP-25A, ... | | Authors: | Ernst, J.A, Brunger, A.T. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High Resolution Structure, Stability, and Synaptotagmin Binding of a Truncated Neuronal SNARE Complex
J.Biol.Chem., 278, 2003
|
|
1DQV
 
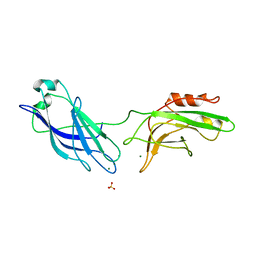 | | CRYSTAL STRUCTURE OF SYNAPTOTAGMIN III C2A/C2B | | Descriptor: | MAGNESIUM ION, SULFATE ION, SYNAPTOTAGMIN III | | Authors: | Sutton, R.B, Ernst, J.A, Brunger, A.T. | | Deposit date: | 2000-01-05 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the cytosolic C2A-C2B domains of synaptotagmin III. Implications for Ca(+2)-independent snare complex interaction.
J.Cell Biol., 147, 1999
|
|
1IHW
 
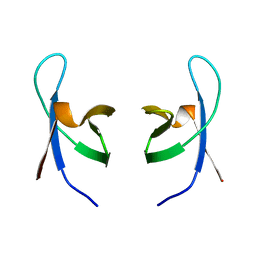 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
1IHV
 
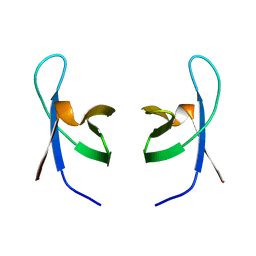 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-10-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
3HN8
 
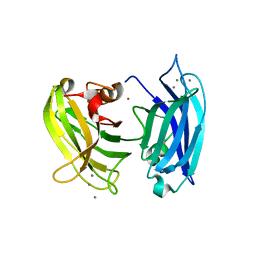 | | Crystal structure of synaptotagmin | | Descriptor: | CALCIUM ION, Synaptotagmin-3, ZINC ION | | Authors: | Strop, P, Vrljic, M, Ernst, J, Brunger, A.T. | | Deposit date: | 2009-05-30 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanism of the synaptotagmin-SNARE interaction in Ca2+-triggered vesicle fusion.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6N9T
 
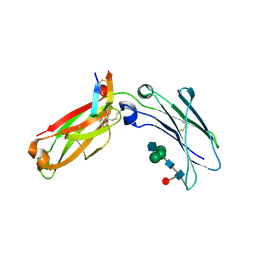 | | Structure of a peptide-based photo-affinity cross-linker with Herceptin Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin G1 FC, ... | | Authors: | Sadowsky, J, Ultsch, M, Vance, N, Wang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | Development, Optimization, and Structural Characterization of an Efficient Peptide-Based Photoaffinity Cross-Linking Reaction for Generation of Homogeneous Conjugates from Wild-Type Antibodies.
Bioconjug. Chem., 30, 2019
|
|
5UQV
 
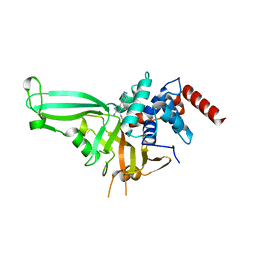 | |
5UQX
 
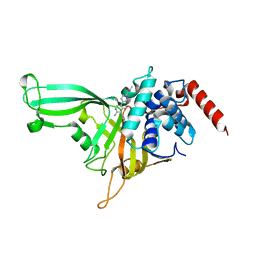 | |
5VZY
 
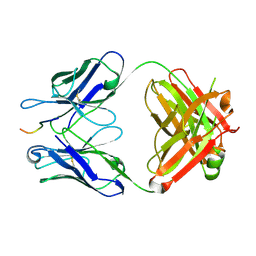 | | Crystal structure of crenezumab Fab in complex with Abeta | | Descriptor: | Amyloid beta A4 protein, Crenezumab Fab heavy chain,Immunoglobulin gamma-1 heavy chain, Crenezumab Fab light chain,Immunoblobulin light chain | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
5VZX
 
 | | Crystal structure of crenezumab Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Crenezumab Fab heavy chain, ... | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
5J7T
 
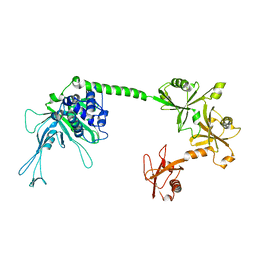 | |
5JTJ
 
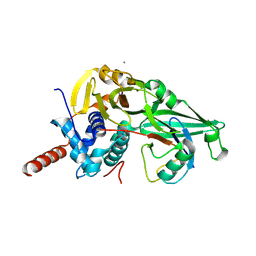 | | USP7CD-CTP in complex with Ubiquitin | | Descriptor: | CALCIUM ION, Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 7,Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation.
Structure, 24, 2016
|
|
5JTV
 
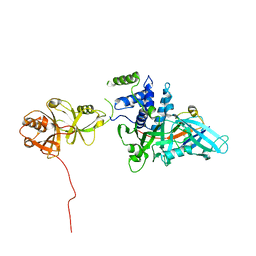 | | USP7CD-UBL45 in complex with Ubiquitin | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation.
Structure, 24, 2016
|
|
