353D
 
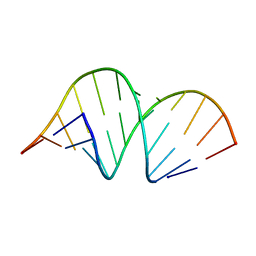 | | CRYSTAL STRUCTURE OF DOMAIN A OF THERMUS FLAVUS 5S RRNA AND THE CONTRIBUTION OF WATER MOLECULES TO ITS STRUCTURE | | Descriptor: | RNA (5'-R(*AP*UP*CP*CP*CP*CP*CP*GP*UP*GP*CP*C)-3'), RNA (5'-R(*GP*GP*UP*GP*CP*GP*GP*GP*GP*GP*AP*U)-3') | | Authors: | Betzel, C, Lorenz, S, Furste, J.P, Bald, R, Zhang, M, Schneider, T.R, Wilson, K.S, Erdmann, V.A. | | Deposit date: | 1997-09-29 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of domain A of Thermus flavus 5S rRNA and the contribution of water molecules to its structure.
FEBS Lett., 351, 1994
|
|
3GVN
 
 | | The 1.2 Angstroem crystal structure of an E.coli tRNASer acceptor stem microhelix reveals two magnesium binding sites | | Descriptor: | 5'-R(*CP*CP*UP*CP*AP*CP*C)-3', 5'-R(*GP*GP*UP*GP*AP*GP*G)-3', MAGNESIUM ION | | Authors: | Eichert, A, Furste, J.P, Schreiber, A, Perbandt, M, Betzel, C, Erdmann, V.A, Forster, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2A crystal structure of an E. coli tRNASer)acceptor stem microhelix reveals two magnesium binding sites.
Biochem.Biophys.Res.Commun., 386, 2009
|
|
361D
 
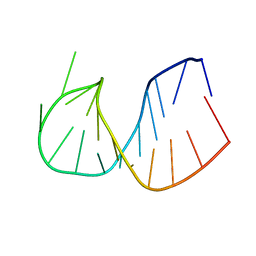 | | CRYSTAL STRUCTURE OF DOMAIN E OF THERMUS FLAVUS 5S RRNA: A HELICAL RNA-STRUCTURE INCLUDING A TETRALOOP | | Descriptor: | RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*GP*GP*CP*GP*AP*CP*CP*GP*CP*C P*UP*GP*G)-3') | | Authors: | Perbandt, M, Nolte, A, Lorenz, S, Erdmann, V.A, Betzel, C. | | Deposit date: | 1997-11-10 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of domain E of Thermus flavus 5S rRNA: a helical RNA structure including a hairpin loop.
FEBS Lett., 429, 1998
|
|
439D
 
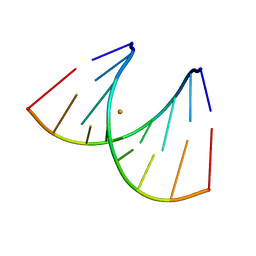 | | 5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3', 5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3' | | Descriptor: | BARIUM ION, RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3') | | Authors: | Perbandt, M, Lorenz, S, Vallazza, M, Erdmann, V.A, Betzel, C. | | Deposit date: | 1999-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an RNA duplex with an unusual G.C pair in wobble-like conformation at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1R3O
 
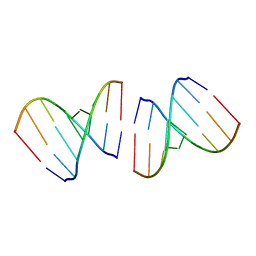 | | Crystal structure of the first RNA duplex in L-conformation at 1.9A resolution | | Descriptor: | L-RNA | | Authors: | Vallazza, M, Perbandt, M, Klussmann, S, Rypniewski, W, Erdmann, V.A, Betzel, C. | | Deposit date: | 2003-10-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First look at RNA in L-configuration.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SDR
 
 | | CRYSTAL STRUCTURE OF AN RNA DODECAMER CONTAINING THE ESCHERICHIA COLI SHINE-DALGARNO SEQUENCE | | Descriptor: | RNA (5'-R(*AP*UP*CP*AP*CP*CP*UP*CP*CP*UP*UP*A)-3'), RNA (5'-R(*UP*AP*AP*GP*GP*AP*GP*GP*UP*GP*AP*U)-3') | | Authors: | Schindelin, H, Zhang, M, Bald, R, Fuerste, J.-P, Erdmann, V.A, Heinemann, U. | | Deposit date: | 1994-12-11 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence.
J.Mol.Biol., 249, 1995
|
|
2G32
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2G5L
 
 | | Streptavidin in complex with Nanotag | | Descriptor: | (FME)(ASP)(VAL)(GLU)(ALA)(TRP)(LEU), GLYCEROL, SULFATE ION, ... | | Authors: | Perbandt, M, Bruns, O, Vallazza, M, Lamla, T, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-23 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High resolution structure of Streptavidin in complex with a novel high affinity peptide tag mimicking the biotin binding motif
To be Published
|
|
2GQ4
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GQ6
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GQ5
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GPM
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3') | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GQ7
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2R9K
 
 | | Crystal Structure of Misteltoe Lectin I in Complex with Phloretamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-hydroxyphenyl)propanamide, ... | | Authors: | Meyer, A, Rypniewski, W, Celewicz, L, Erdmann, V.A, Voelter, W, Betzel, C. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mistletoe lectin I--phloretamide structure reveals a new function of plant lectins.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
2V7R
 
 | | Crystal structure of a human tRNAGly microhelix at 1.2 Angstrom resolution | | Descriptor: | HUMAN TRNAGLY MICROHELIX | | Authors: | Foerster, C, Mankowska, M, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-08-01 | | Release date: | 2008-03-18 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a Human Trnagly Microhelix at 1.2 A Resolution.
Biochem.Biophys.Res.Commun., 368, 2008
|
|
2V6W
 
 | | tRNASer acceptor stem: Conformation and hydration of a microhelix in a crystal structure at 1.8 Angstrom resolution | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*AP)-3', 5'-R(*UP*CP*UP*CP*UP*CP*CP)-3' | | Authors: | Foerster, C, Brauer, A.B.E, Brode, S, Fuerste, J.P, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-11-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trnaser Acceptor Stem: Conformation and Hydration of a Microhelix in a Crystal Structure at 1.8 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2VAL
 
 | | Crystal structure of an Escherichia coli tRNAGly microhelix at 2.0 Angstrom resolution | | Descriptor: | 5'-R(*GP*CP*GP*GP*GP*AP*AP)-3', 5'-R(*UP*UP*CP*CP*CP*GP*CP)-3', MAGNESIUM ION | | Authors: | Forster, C, Brauer, A.B.E, Perbandt, M, Lehmann, D, Furste, J.P, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of an Escherichia Coli Trnagly Microhelix at 2.0 Angstrom Resolution
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2W89
 
 | | Crystal structure of the E.coli tRNAArg aminoacyl stem issoacceptor RR-1660 at 2.0 Angstroem resolution | | Descriptor: | 5'-R(*CP*GP*GP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*CP*CP*GP)-3', GLYCEROL | | Authors: | Eichert, A, Schreiber, A, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E. Coli tRNA(Arg) Aminoacyl Stem Isoacceptor Rr-1660 at 2.0 A Resolution.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
2VUQ
 
 | | Crystal structure of a human tRNAGly acceptor stem microhelix (derived from the gene sequence DG9990) at 1.18 Angstroem resolution | | Descriptor: | 5'-R(*CP*CP*AP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*UP*GP*GP)-3' | | Authors: | Eichert, A, Perbandt, M, Schreiber, A, Fuerste, J.P, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2008-05-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal Structure of the Human Trnagly Microhelix Isoacceptor G9990 at 1.18 A Resolution
Biochem.Biophys.Res.Commun., 380, 2009
|
|
2X2Q
 
 | | Crystal structure of an 'all locked' LNA duplex at 1.9 angstrom resolution | | Descriptor: | CACODYLATE ION, COBALT HEXAMMINE(III), LOCKED NUCLEIC ACID DERIVED FROM TRNA SER ACCEPTOR STEM MICROHELIX, ... | | Authors: | Eichert, A, Behling, K, Fuerste, J.P, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2010-01-15 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of an 'All Locked' Nucleic Acid Duplex.
Nucleic Acids Res., 38, 2010
|
|
2XSL
 
 | | The crystal structure of a Thermus thermophilus tRNAGly acceptor stem microhelix at 1.6 Angstroem resolution | | Descriptor: | 5'-R(*CP*UP*CP*CP*CP*GP*C)-3', 5'-R(*GP*CP*GP*GP*GP*AP*G)-3' | | Authors: | Oberthuer, D, Eichert, A, Erdmann, V.A, Fuerste, J.P, Betzel, C, Foerster, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-08-10 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Crystal Structure of a Thermus Thermophilus tRNA(Gly) Acceptor Stem Microhelix at 1.6 A Resolution.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
